Fig. 9.
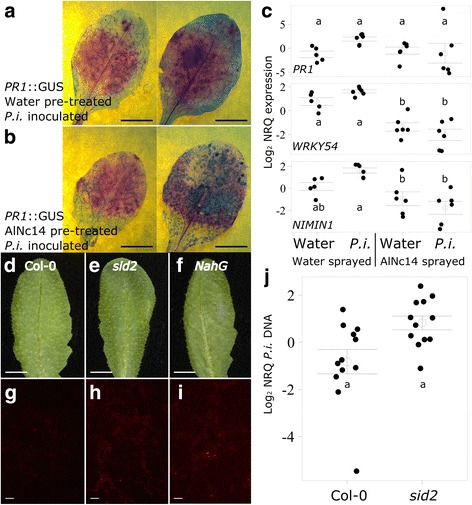
Albugo suppression of Arabidopsis salicylic acid (SA) responses is not sufficient for full susceptibility to P. infestans. a and b PR1::GUS staining upon P. infestans infection. Leaves were collected and stained with magenta-GUS to reveal GUS activity, followed by trypan blue to reveal pathogen growth. PR1::GUS plants were pre-treated with water or AlNc14 and subsequently inoculated with 100 μL of 1.25 × 105 spores per mL P. infestans 88069td, collected at 2 dpi and stained. Scale: 5 mm. Representative leaves shown are from each of two independent experiments. c AlNc14 infection prevents P. infestans-induced upregulation of SA marker genes in Col-0. Open circles and bars denote the mean ± SE of target gene expression (log2 normalized relative quantities (NRQs)) at 48 hours post treatment (100 μL water or P. infestans (1.25 × 105 spores per mL)) of three independent biological replicates with two technical replicates each. Closed, black circles denote the individual data points. Different letters indicate significant differences (P < 0.05; two-way ANOVA with Tukey’s HSD test). d–i P. infestans partially colonizes sid2 and NahG Arabidopsis. Leaves were inoculated with 100 μL of 1 × 105 spores per mL P. infestans 88069td, photographed (d–f) and the adaxial surface examined using fluorescence microscopy (g–i) at 3 dpi. Red fluorescence denotes P. infestans growth, Scale bars: 5 mm for photographs, 1 mm for microscopy. Results shown are representative of three independent experiments. j P. infestans growth on sid2 is not significantly larger than Col-0 Arabidopsis. Leaves were inoculated as in d, e, g, h. DNA was extracted at 3 dpi and the proportion of P. infestans DNA to plant DNA determined using qRT-PCR. Open circles and bars denote the mean ± SE of P. infestans DNA (log2 transformed NRQs) in Arabidopsis tissue from four independent biological replicates with three technical replicates each. Closed, black circles denote the individual data points. The two genotypes were not significantly different (P = 0.012) (Wilcoxon rank sum test followed by Bonferroni correction)
