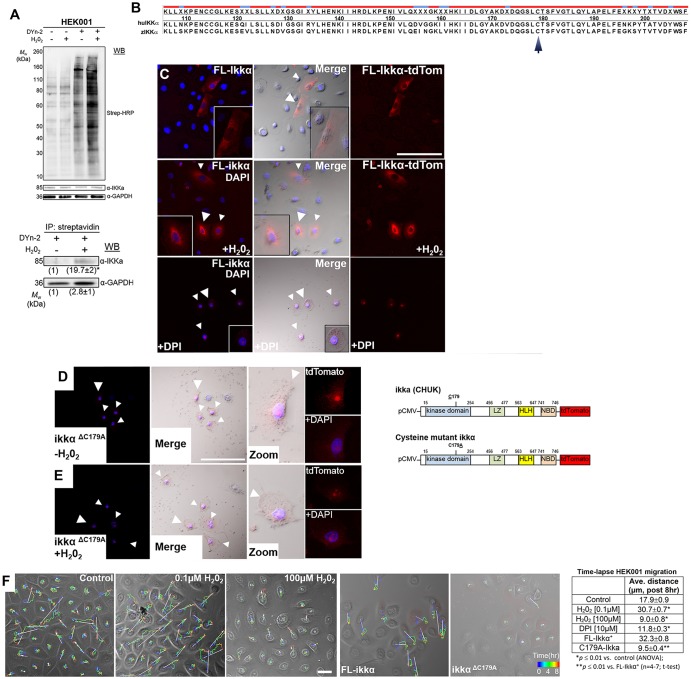
Fig. 5.
IKKα sulfenylation promotes keratinocyte migration. (A) Upper blot: DYn-2 probe-loaded keratinocytes were biotinylated using ‘click chemistry’ with a biotinylated azide probe. Sulfenylated proteins were probed with streptavidin (Strep)–HRP (upper panel). IKKα is detected in all fractions (with or without DYn-2 and H2O2). Lower blot: H2O2 oxidizes IKKα, assessed after purification of the biotinylated fraction by streptavidin pull-down and probing with anti-IKKα antibody. GAPDH serves as positive control for successful pull down of oxidized/sulfenylated proteins given its redox function (Truong, 2012a). IP, immunoprecipitation; WB, western blot. Values under the blots depict fold-changes from baseline controls. (B) Human (Hu) and zebrafish (z) IKKα kinase domain sequence comparison shows high conservation (∼70%), including zebrafish residue cysteine 179 (arrow). Red, conserved residues; blue, non-conserved residues. (C) IKKα construct scheme for HEK001 in vitro transfection studies are shown to the right of panel D. Full-Length (FL) Ikkα–tdTomato in cytoplasmic perinuclear regions with and without H2O2 (white arrowheads). Large arrowheads indicate cells magnified in the insets. DPI treatment (10 µM) results in punctate localization within subregions of the nucleus. Scale bar: 100 µm. (D,E) Point mutant (C179A) shows nuclear localization without H2O2 (D) and with H2O2 (0.1 µM) (E). Scale bar: 100 µm. Arrowheads, indicate transfected cells; large arrowheads indicated cell magnified in the insets. (F) Cell movement and directionality (arrows) tracked over time. Control (untreated) cells migrate slightly faster than cells treated with high H2O2 (100 µM) concentrations, following DPI treatment and when transfected with C179A-Ikk. Transfection with FL-Ikk and treatment with low H2O2 (0.1 µM) concentrations promotes migration. Scale bar: 40 µm. Ave, average, mean; LZ, leucine zipper; HLH, helix-loop-helix; NBD, Nemo-binding domain. Data are mean±s.d. ANOVA test and Tukey's multiple comparisons test were used for statistical analyses.