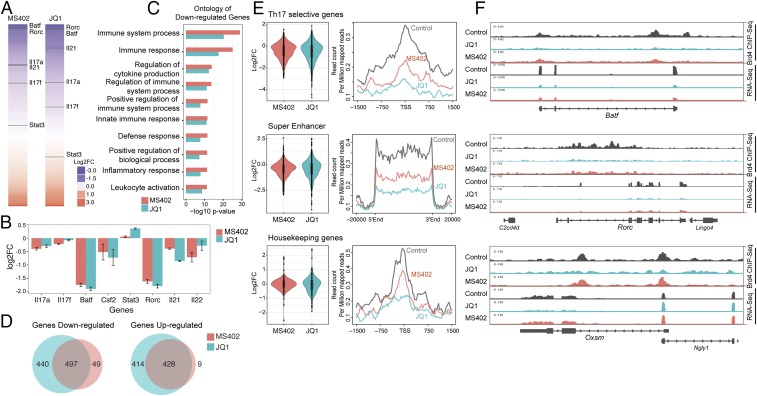
Fig. 3.
Genomic analysis of BET inhibition effects on gene transcription during Th17 cell differentiation. (A) Effects of MS402 and JQ1 treatment on gene transcription in Th17 cell differentiation. Genes are ranked from most down-regulated to most up-regulated by compound treatment. Select Th17 signature genes whose transcriptional levels are affected by compound treatment are indicated. (B) MS402 and JQ1 show comparable (log2) fold change for select Th17 signature genes. (C) Immune and cytokine ontologies are among top-enriched down-regulated genes by MS402 or JQ1 treatment. (D) Venn diagram analysis of genes down- or up-regulated by MS402 and JQ1 treatment during Th17 differentiation. (E) Violin plots of the RNA-seq data and profiles of the ChIP-seq data showing patterns of perturbation across multiple gene sets of Th17 selective genes, genes with super enhancers, and housekeeping genes in Th17 cells upon treatment of MS402 or JQ1. (F) Select RNA-seq tracks illustrating changes of transcriptional expression of Brd4 target genes Batf and Rorc (Th17 selective genes) and Oxsm (housekeeping) upon the treatment of MS402 or JQ1.