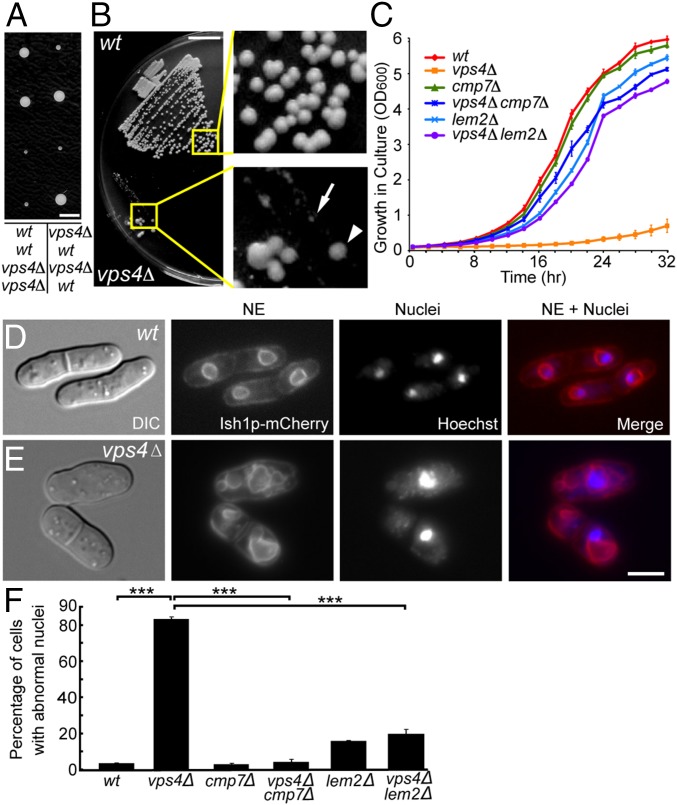
Fig. 1.
vps4Δ cells grow slowly and have severe NE defects, which are suppressed by loss of cmp7 or lem2. (A) Tetrad dissection of vps4Δ/+ diploids, with genotypes labeled below the image. (Scale bar: 0.25 cm.) (B) Spontaneous suppressors (arrowhead) of vps4Δ (arrow) appearing after 3 d on rich medium, with colony sizes comparable to those of WT cells. (Scale bar: 1 cm.) (C) Growth curves of each genotype showing optical densities of yeast cultures, starting at OD600nm 0.06, for 32 h in 2-h intervals. Three independent isolates for each genotype were measured. The plots show mean ± SEM. (D) WT cells, showing normal NE morphologies (Ish1p-mCherry) and DNA (Hoechst staining) within the nuclei. (E) vps4Δ, showing various NE morphology defects including excess NE, fragmented NE, and DNA that appears to be outside of the nucleus. (Scale bar: 5 μm.) (F) Deletion of either cmp7 or lem2 rescues these NE morphology defects. Here and throughout, three independent isolates/strains were imaged for each genotype, and n represents the total number of scored cells. Mean ± SEM, WT: 1 ± 1%, n = 115, 121, 139; vps4Δ: 85 ± 5%, n = 62, 69, 86; cmp7Δ: 2.0 ± 0.4%, n = 151, 166, 147; vps4Δcmp7Δ: 1.1 ± 0.7%, n =161, 118, 145; lem2Δ: 8 ± 3%, n= 131, 103, 128; vps4Δlem2Δ: 12.2 ± 0.7%, n = 99, 176, 111. Two-tailed Student t tests were used here and throughout. ***P < 0.001.