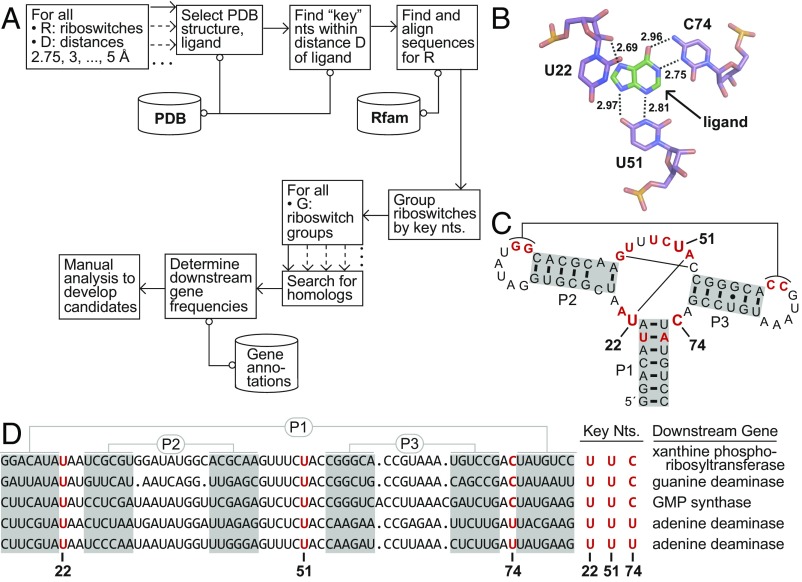
Fig. 1.
Bioinformatic search method illustrated using guanine and adenine riboswitch examples. (A) Schematic depiction of a process to detect riboswitches with altered ligand specificities (see text for details). (B) Atomic-resolution model of the ligand-binding pocket of a guanine riboswitch aptamer bound to the guanine analog hypoxanthine (26) (PDB ID code 4ef5). Three “key” nucleotides (U22, U51, and C74) of the aptamer carry atoms that are within 3 Å of a ligand atom (see dashed lines). The same three key nucleotides would have been identified if the natural ligand guanine were docked. (C) Key nucleotides at positions 22, 51, and 74 mapped onto the sequence and secondary-structure model for the guanine riboswitch aptamer whose X-ray structure was used to conduct this analysis (26). Nucleotides in red identify positions that are conserved in 97% or greater of the known guanine riboswitch aptamers. Thin lines identify long-range base pairs. (D) Alignment of the sequence of the guanine riboswitch aptamer in B and C with two additional guanine and adenine aptamers, arranged from Top to Bottom, respectively. Adenine riboswitches, known to carry a C-to-U mutation at position 74, commonly regulate adenine deaminase genes that are not regulated by these three guanine riboswitches. The complete analysis for this collection of riboswitches included 3,462 guanine and 187 adenine riboswitches.