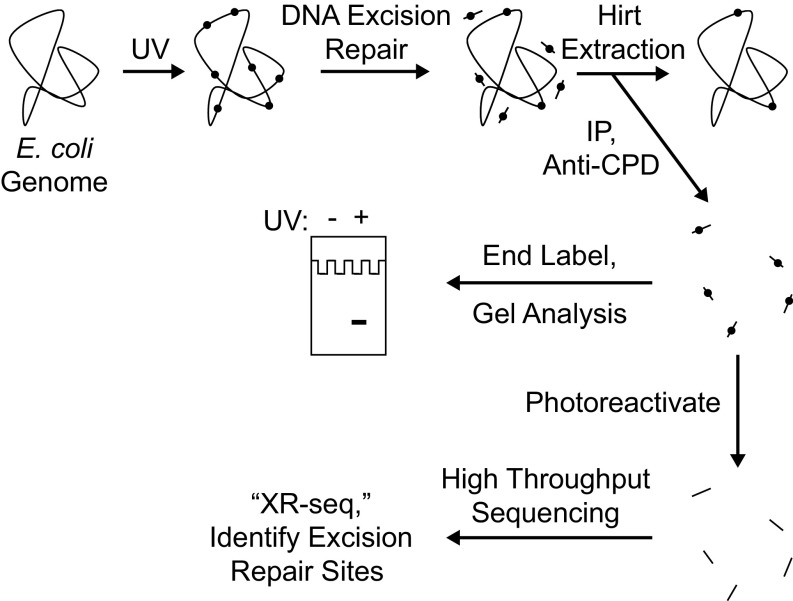
Fig. 1.
Scheme for analyzing repair of the E. coli genome. Cells are irradiated with UVC (254 nm) and then incubated for a period to allow nucleotide excision repair. Cells are then harvested and cooled on ice, and then the Hirt procedure (23) is used to separate relatively small DNA molecules from genomic DNA and cellular debris. From the preparation of small DNA molecules, DNA molecules containing cyclobutane pyrimidine dimers (CPDs) are then isolated by immunoprecipitation with an anti-CPD–specific antibody. The isolated products may be analyzed in two ways. To detect the overall excision of damage from the genome, samples may be directly end-labeled with 32P and subjected to denaturing polyacrylamide gel electrophoresis. To map the sites of DNA excision repair throughout the genome at nucleotide resolution, products may be analyzed by high-throughput sequencing after they are ligated to adaptors, immunoprecipitated again with anti-CPD antibody, photoreactivated, amplified by PCR, and gel-purified (25).