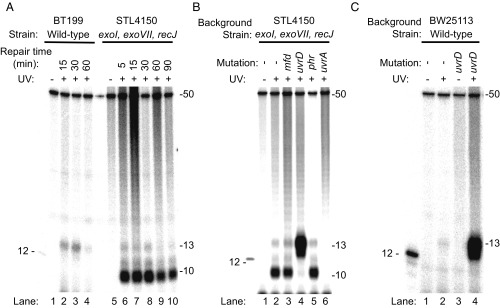
Fig. 2.
Analysis of nucleotide excision repair products from E. coli by excision assay. Cells were irradiated with 100 J/m2 (A and B) or 120 J/m2 (C) and then incubated for the indicated times (in A) or for 5 min (B and C). Repair products containing a CPD were isolated from cells, end-labeled with 32P, and resolved by denaturing polyacrylamide gel electrophoresis. The gel images show a predominantly 13-mer size excision product obtained from WT cells, which is consistent with results from in vitro analysis (27). This 13-mer size excision product is simultaneously generated and degraded in vivo, and a reduction in the amount of the 13-mer product seen after 30 min is consistent with an ∼30-min time course to complete nucleotide excision repair in E. coli cells (3). Strain STL4150 lacks the major ssDNA exonucleases, and the images in A and B show that, in STL4150 cells, there is limited degradation of the 13-mer past the 10-mer stage. In uvrD mutant cells (B and C), there is an elevated level of 13-mer product. The UvrD protein is the major helicase in E. coli. In nucleotide excision repair, UvrD catalyzes the displacement of the damage-containing 13-mer excision product and initiates displacement of the UvrB and UvrC proteins from the genome. Consequently, in these cells, the 13-mer remains annealed to the genome where it is resistant to nucleases. This accumulation of the 13-mer is observed even though uvrD mutant cells excise the 13-mer slowly because turnover of UvrB and UvrC is slow. The 12-mer marker DNA shown in the images contains a CPD and was end-labeled with polynucleotide kinase. The 50-mer (1 fmol) was included in each sample before end-labeling, as an internal control.