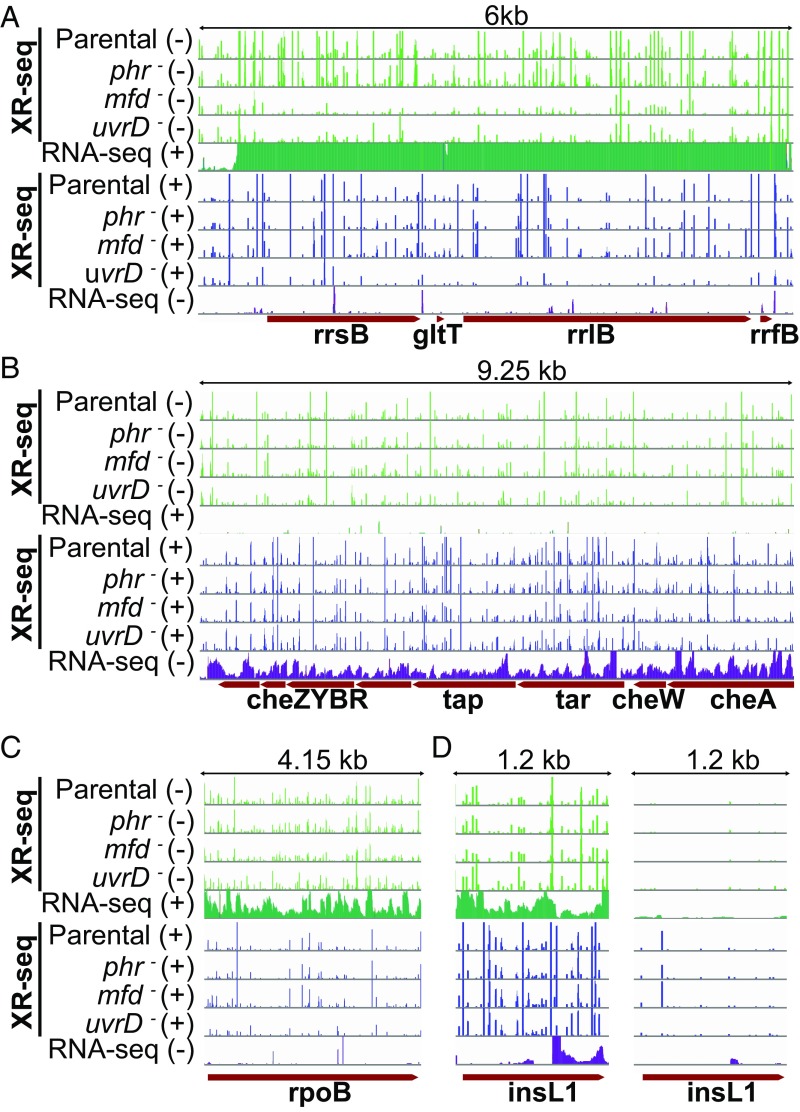
Fig. 6.
XR-seq and RNA-seq patterns of exemplified operons and genes. In all panels, the first four green-colored rows represent the XR-seq reads of the four strains studied aligned with the minus strand of the reference genome (E. coli K-12 MG1655 genome with NCBI accession NC_000913.2). The fifth green bar shows the RNA-seq reads aligned with the plus strand. Blue bars represent the opposite strand. For the genes that are on the plus strand (A, C, and D), the green XR-seq reads correspond to the transcribed strand repair where green RNA-seq reads represent the RNA products that are due to the sense transcription. Blue XR-seq data illustrate repair of the coding (plus) strand, and blue RNA-seq reads represent the antisense transcription. The opposite is true for the genes on the minus strand (B). The y axis is scaled to show 105 counts for each bar except for the right panel of D, where the y axis is scaled up to 2,000. (A) The illustrated 6-kb genomic window is between 4,164 and 4,170 kb, which is an rRNA operon rrnB with 16S (rrsB), 23S (rrlB), and 5S (rrfB) rRNA genes, as well as the glutamate tRNA (gltT) gene. This operon was selected to represent a high level of transcription so that the Mfd effect is drastically visible. (B) Chemotaxis operon (1964.25 to 1973.5 kb) with moderate level of transcription. (C) The rpoB gene (4179.2 to 4183.35 kb) displays an amplified repair signal in the uvrD strain. (D) The insL1 gene (2512.3 to 2513.5 kb) with antisense transcription in the 3′ end (Left).