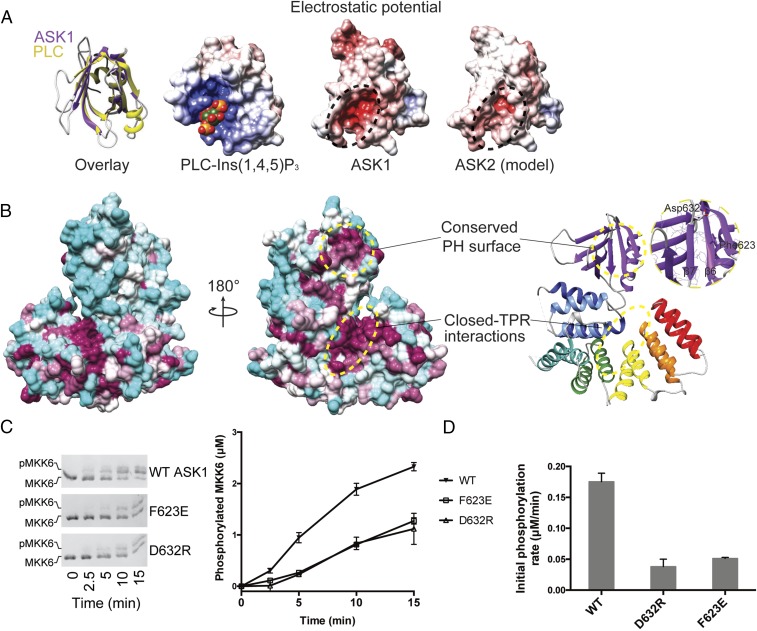
Fig. 3.
The ASK1 pleckstrin homology domain mediates protein-protein interactions and activity. (A, Left) Superposition of the ASK1 pleckstrin homology domain (purple) with the canonical pleckstrin homology domain of phospholipase C (PLC, in yellow; PDB ID code 1MAI) (77). (A, Right) Three electrostatic surface representations calculated in APBS (78) for the pleckstrin homology domains of PLC bound to Ins(1,4,5)P3, ASK1, and a model of the same region of ASK2 generated using MODELER. (Conservation between ASK1 and ASK2 can be viewed in the alignment in Fig. S4.) (B) Surface representation of ASK1(269–658), color-coded according to the degree of conservation in the alignment in Fig. S4. The least conserved residues are in cyan; the most conserved, in maroon. Areas of high conservation are also indicated with circles. (Inset) Close-up view of the conserved pleckstrin homology surface. (C) Phos-tag SDS/PAGE monitoring MKK6 (3 μM) phosphorylation by ASK1(88–941) (1 μM) WT or indicated pleckstrin homology domain mutants. Quantitation of independent triplicate experiments is shown alongside as mean values, with error bars representing SEM. (D) Rates of MKK6 phosphorylation calculated over the first 5 min from C. Error bars represent the SD of the linear rate fit.