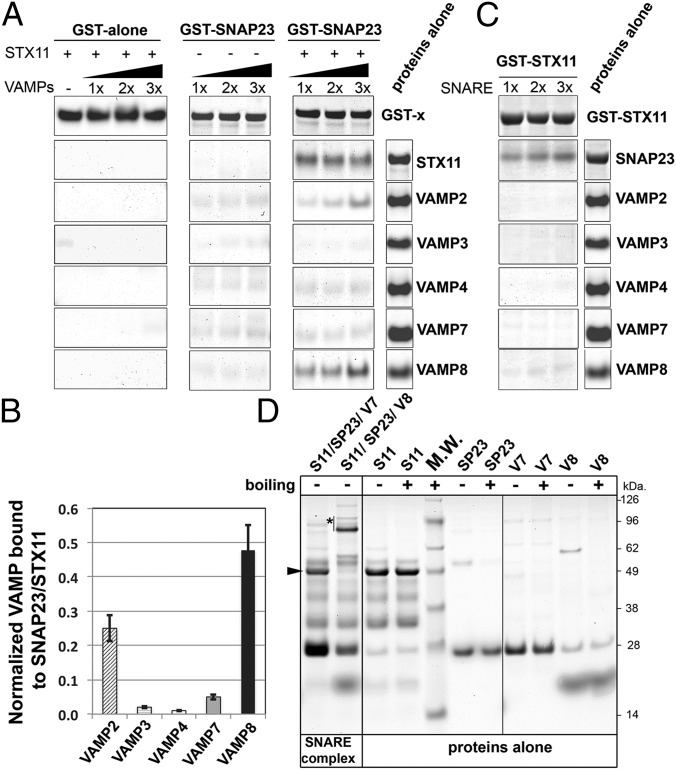
Fig. 4.
VAMP8 or VAMP2 bind to a recombinant STX11/GST–SNAP23 complex. (A) Equivalent amounts of recombinant GST (GST alone) or GST–SNAP23 were bound to glutathione-Sepharose beads, and increasing concentrations (0.5, 1.0, or 1.5 μg) of recombinant VAMP2, -3, -4, -7, and -8 were added to the beads in the absence or presence of 1.0 μg recombinant STX11 (amino acids 1–276) and incubated for 1 h at room temperature. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Proteins-alone lane shows representative images of the total load of each protein used in these experiments. (B) Plot showing quantification of the amount of VAMP protein bound to the beads normalized to the amount of STX11 retained in each experiment. Data represent mean ± SD of three independent experiments. (C) Recombinant GST–STX11 was bound to glutathione-Sepharose beads, and increasing concentrations (0.5, 1.0, or 1.5 μg) of recombinant SNAP23 or VAMP2, -3, -4, -7, and -8 were added to the beads. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Proteins-alone lane is identical to the one shown in A because both experiments were performed in parallel with the same batch of proteins. (D) Recombinant His–SUMO–STX11 was incubated with SNAP23 and either VAMP8 or VAMP7 for 1 h at room temperature, and the formation of high molecular weight complexes was analyzed by SDS/PAGE without boiling the samples. Arrowhead shows the band corresponding to His–SUMO–STX11, which largely disappeared upon SNARE complex formation with SNAP23 and VAMP8 (asterisk), but not with VAMP7. Proteins-alone lane shows the load of each protein used to generate the SNARE complexes. M.W., molecular weight markers (shown to the Right of the gel).