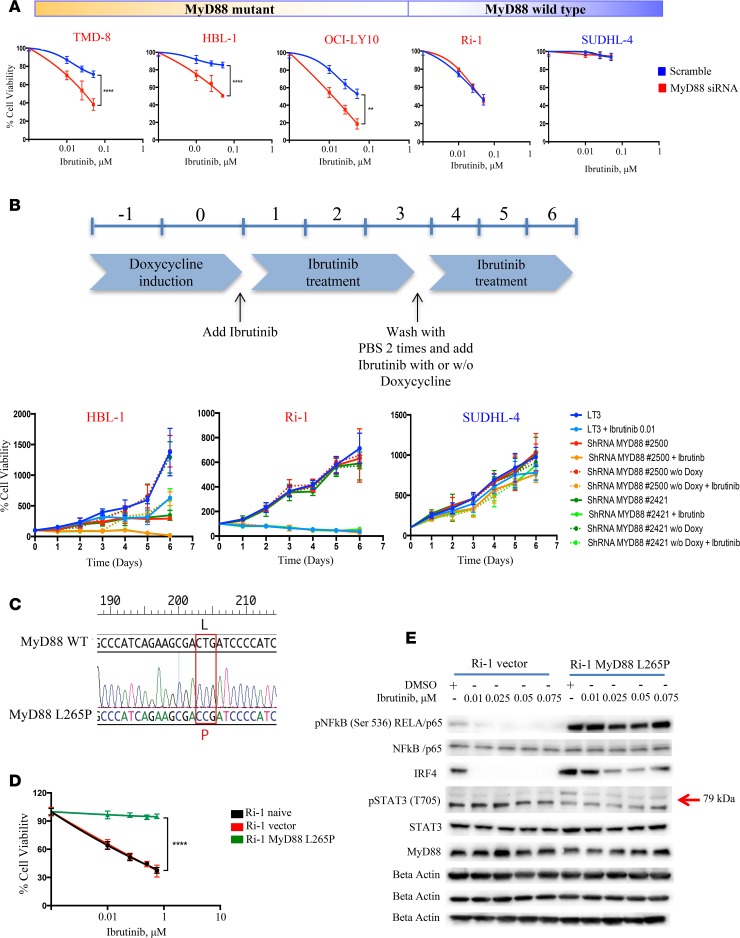
Figure 1. MyD88 confers resistance to ibrutinib in ABC DLBCL.
(A) Five DLBCL cells (3 ABC MyD88 mutant [TMD-8, HBL-1, and OCI-LY10], 1 ABC MyD88 WT [Ri-1] and 1 GCB [SUDHL-4]) were treated with 1 μM scramble, and MyD88 siRNA were incubated with increasing concentrations of ibrutinib (0.1, 0.25, 0.5 μM) and cell viability assessed by MTS assay after 24 hours. MyD88 siRNA + ibrutinib viability data were normalized to the effect of MyD88 siRNA alone. Error bars represent SEM of triplicate experiments. Differences between groups were calculated with the Student t test. **P < 0.005; ****P < 0.0001. (B) HBL-1, Ri-1 (ABC MyD88 mutant and WT, respectively) and SUDHL-4 (GCB) cells were infected with either tet inducible control shRNA or shRNA against MyD88, selected with puromycin, and induced with doxycycline for 2 days before treating with ibrutinib and seeding for MTS assay. After 3 days from the beginning of drug treatment, doxycyxline was washed out and ibrutinib added fresh either with or without fresh doxycycline. Error bars represent SEM of triplicate experiments. (C) Analyses of DNA sequences of a MyD88 WT and MyD88 L265P mutation, showing the replacement of reference sequence CTG with CCG. (D) Ri-1 naive cells transduced with either empty vector or activated MyD88 mutant L265P were incubated with increasing concentrations of ibrutinib (0.1, 0.25, 0.5, 0.75 μM) and cell viability assessed by MTS assay after 24 hours. MyD88 mutant + ibrutinib viability data were normalized to the effect of MyD88 mutant alone. Error bars represent SEM of triplicate experiments. Differences between groups were calculated with the Student t test. ****P < 0.0001. (E) Representative Western blot demonstrating inhibition of NF-κB pathway (pNF-κB p65, IRF4, pSTAT3 (T705]) after treatment with increasing concentrations of ibrutinib (0.1, 0.25, 0.5, 0.75 μM) for 24 hours in Ri-1 MyD88 WT but not in the Ri-1 stably expressing MyD88 mutation.