Abstract
Women diagnosed with breast cancer within 5 years of childbirth have poorer prognosis than nulliparous or pregnant women. Weaning-induced breast involution is implicated, as the collagen-rich, immunosuppressive microenvironment of the involuting mammary gland is tumor promotional in mice. To investigate the role of mammary fibroblasts, isolated mammary PDGFRα+ cells from nulliparous and postweaning mice were assessed for activation phenotype and protumorigenic function. Fibroblast activation during involution was evident by increased expression of fibrillar collagens, lysyl oxidase, Tgfb1, and Cxcl12 genes. The ability of mammary tumors to grow in an isogenic, orthotopic transplant model was increased when tumor cells were coinjected with involution-derived compared with nulliparous-derived mammary fibroblasts. Mammary tumors in the involution-fibroblast group had increased Ly6C+ monocytes at the tumor border, and decreased CD8+ T cell infiltration and tumor cell death. Ibuprofen treatment suppressed involution-fibroblast activation and tumor promotional capacity, concurrent with decreases in tumor Ly6C+ monocytes, and increases in intratumoral CD8+ T cell infiltration, granzyme levels, and tumor cell death. In total, our data identify a COX/prostaglandin E2 (PGE2)–dependent activated mammary fibroblast within the involuting mammary gland that displays protumorigenic, immunosuppressive activity, identifying fibroblasts as potential targets for the prevention and treatment of postpartum breast cancer.
Introduction
A diagnosis of breast cancer within 5 to 10 years of childbirth is an independent predictor of poor prognosis owing to increased risk of metastasis (1–4). Further, an estimated 40% to 50% of all young women’s breast cancers are associated with a recent pregnancy, identifying postpartum breast cancer as a significant unmet clinical need (3). Unexpectedly, the poor outcomes of postpartum breast cancer do not appear to be due to pregnancy per say, as the prognosis of breast cancers diagnosed during pregnancy is not significantly worse than breast cancers diagnosed in nulliparous women (1, 2). Rather, data implicate a unique biology of the postpartum breast in the promotion of postpartum breast cancer. Specifically, weaning-induced mammary gland involution is implicated because, in rodents, the tissue microenvironment of the involuting mammary gland enhances tumor growth, local tumor cell dissemination, and distant metastasis (5–7).
Weaning-induced mammary gland involution is a biologically normal process, whereby milk stasis induces gland regression to a nonlactational state. Mammary gland involution is well characterized in rodents (8–12) and recently described in women (12, 13). Involution can be separated broadly into an initial phase of secretory epithelial cell death followed by a stromal remodeling phase that reestablishes the adipocytes and connective tissue as dominant constituents of the nonlactating gland (8, 14). The stromal remodeling phase of involution shares numerous attributes with wound healing (15–17), including lymphangiogenesis (6), orchestrated immune cell infiltration, immune suppression (7, 12), and deposition of a fibrotic-like extracellular matrix (ECM), mainly the fibrillar collagens (9, 12). The mechanistic links between wound healing and cancer (18, 19) and the known role of fibroblasts in these processes (20–23) identify the mammary fibroblast as a potential mediator of postpartum breast cancer progression.
Fibrillar collagen deposition, which is increased during mammary gland involution, is one mechanism by which mammary fibroblasts might promote breast cancer in the postpartum window (12). Fibrillar collagens are the most abundant ECM proteins in the breast and contribute to breast density, a dominant risk factor for breast cancer in women (24, 25). Fibrillar collagens are also implicated in human breast cancer progression, as the presence of straight collagen fibers arranged perpendicular to the tumor border, an orientation referred to as radial alignment, is an independent predictor of breast cancer metastasis (26). Further, in preclinical models of breast cancer, collagen density and tension regulate progression of preexisting tumor cells (27–29). Intravital imaging studies provide additional support for collagen mediating tumor cell dissemination, as tumor cells migrate on mammary collagen fibers to gain access to vasculature (30, 31). We have reported that targeting the window of postpartum involution with nonsteroidal antiinflammatory agents (NSAIDs) reduces the levels of the COX2 enzymatic product PGE2 during mammary gland involution (32) and is sufficient to reduce mammary tumor growth and metastasis to levels observed in nulliparous mice (5, 6). The tested NSAIDs in those studies include COX2-nonspecific inhibitors ibuprofen and aspirin and the COX2-specific inhibitor celecoxib. Surprisingly, NSAID-induced tumor protection was accompanied by decreases in mammary fibrillar collagen accumulation during involution (5), implicating collagen as a potential driver of postpartum breast cancers. Cumulatively, these studies raise the possibility that fibroblasts mediate the collagen-rich, protumorigenic microenvironment of the involuting mammary gland through a COX2/PGE2–dependent mechanism.
Immune suppression, which also is increased during normal weaning-induced mammary gland involution (7), is another mechanism by which mammary fibroblasts may contribute to breast cancer progression in the postpartum window. Immune suppression is implicated in breast cancer progression overall (33–35), and analyses of breast cancer patient cohorts show that low CD8+ T cell tumor infiltration, the dominant antitumor cytotoxic T cell (36, 37), is associated with decreased patient survival rate (37–39). In addition to direct suppression by tumor cells, CD8+ T cells can be inhibited by other immune cells within the tumor microenvironment, including immunosuppressive monocytes, tumor-associated macrophages (TAMs), and dendritic cells (40–42), resulting in tumor cell escape from immune surveillance. Of potential relevance, cancer-associated fibroblasts (CAFs) have been reported to regulate immune cell infiltration and immune suppression via secretion of IL-6, TGF-β, and CXCL12 (23, 43, 44). These CAF studies raise the provocative question of whether fibroblasts under physiologic conditions, such as weaning-induced mammary gland involution, contribute to breast cancer by regulating immune cell recruitment and/or immune cell suppression.
In this report, we provide evidence that normal PDGFRα+ cells isolated from involuting mammary glands, referred to as involution-fibroblasts, have a unique activation state, including increased fibrillar collagen, matrix remodeling, and immunomodulatory gene expression patterns. Further, we demonstrate that in comparison with mammary PDGFRα+ cells isolated from nulliparous mice (nulliparous-fibroblasts), involution-fibroblasts promote tumor growth, produce CXCL12, induce monocyte recruitment, and associate with blockade of CD8+ T cell tumor infiltration, identifying a role for fibroblasts in inhibiting intratumoral CD8+ T cell infiltration and tumor cell death. Importantly, we find that the NSAID ibuprofen suppresses these tumor promotional fibroblast attributes. In summary, our studies identify the involution-fibroblast as a unique mediator of murine mammary cancer progression in the postpartum setting, as well as a direct therapeutic target of NSAIDs. These data suggest that further investigation into fibroblast-based strategies for the prevention and treatment of postpartum breast cancer is warranted.
Results
Mammary involution-fibroblasts are uniquely activated.
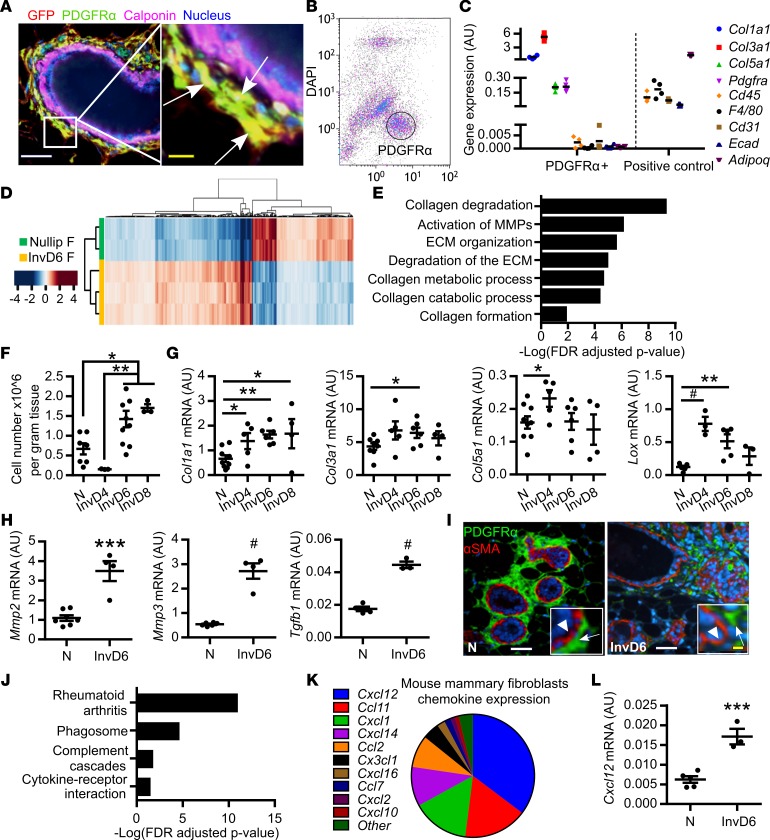
In rodents, mammary stromal remodeling is most active between 4 and 8 days after weaning (8, 12) and previous studies have shown fibrillar collagen protein abundance increases during this window of involution (5, 12). However, the cell type responsible for mammary collagen production has not been definitively described. To this end, we utilized a transgenic, collagen 1a1 reporter mouse in which GFP is driven by the Col1a1 promoter (45). Using dual immunofluorescence, we found GFP-producing cells colocalized with cells expressing the established fibroblast marker PDGFRα (Figure 1A, arrows) (46). These dual-positive cells were found in high abundance surrounding the mammary ducts, but did not colocalize with calponin-positive myoepithelial cells (Figure 1A) (47), ruling out the myoepithelial cell as a dominant cellular source of mammary collagen. IHC analysis demonstrated that ~85% of PDGFRα+ cells were located around the mammary ductal and alveolar structures, with ~15% of staining around the less prevalent blood vessels (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.89206DS1). We next isolated PDGFRα+ cells by positive selection using FACS (Figure 1B). Isolated PDGFRα+ cells were found to express the fibrillar collagen genes Col1a1, Col3a1, and Col5a1, which are the most abundant fibrillar collagens in the rodent mammary gland (48, 49), with low level or no expression of markers for leukocytes (Cd45), macrophages (F4/80), endothelial cells (Cd31), epithelial cells (Ecad), or adipocytes (Adipoq) (Figure 1C). These PDGFRα+ cells were also PDGFRβ+ (Supplemental Figure 1D). Further, the PDGFRα– cell population expressed low levels of fibrillar collagen and lysyl oxidase (Lox) genes compared with the PDGFRα+ cells (Supplemental Figure 1E). Together, these data suggest that PDGFRα+ selection results in a highly enriched mammary fibroblast population.
Figure 1. Mammary fibroblasts are activated during weaning-induced gland involution.
(A) Mammary gland thin sections from the Col1a1-GFP reporter mouse immunofluorescence stained for GFP+ collagen 1–expressing cells (red), fibroblast PDGFRα (green), and myoepithelial cell calponin (purple), show colocalization of GFP and PDGFRα (yellow), but not calponin. Nuclei stained with DAPI (blue). White scale bar: 50 μm. Yellow scale bar: 10 μm. Arrows show PDGFRα+GFP+calponin– cells. (B) Flow gate for isolating murine mammary PDGFRα+ cells. (C) RT-qPCR on FACS-isolated PDGFRα+ cells for fibroblast markers (Col1a1, Col3a1, Col5a1, and PDGFRα) and markers of potential contaminating cell populations: Cd45 (lymphocyte common antigen), F4/80 (mature macrophage), Cd31 (endothelial cell), Adipoq (adipocyte), and Ecad (epithelial cell). Sorted PDGFRα-negative cells were used as positive controls for Cd45, F4/80, Cd31, Adipoq, and mammary epithelial EpH4 cells were used as the Ecad-positive control. n = 3–4 per group. (D) RNA-Seq heatmap illustrates 870 significantly differentially expressed genes (>1.3-fold changes) between fibroblasts isolated from nulliparous and involution day 6 (InvD6) mammary glands. Red: relative high expression. Blue: relative low expression. (E) Pathway analysis of RNA-Seq data set showing extracellular matrix (ECM) regulation–related pathways that are upregulated in InvD6-fibroblasts. (F) PDGFRα+ fibroblasts reported as number per gram of mammary tissue observed in nulliparous and 4-, 6-, and 8-day postweaning mice; n = 3–10 per group. (G) Fibrillar collagens and Lox gene expression analysis by RT-qPCR in sorted PDGFRα+ mammary fibroblasts from nulliparous and 4-, 6-, and 8-day postweaning mice; n = 3–10 per group. (H) Mmp2, Mmp3, and Tgfb1 gene expression analysis by RT-qPCR in nulliparous and InvD6 fibroblasts, n = 6–9 per group. (I) Immunofluorescence staining shows non-colocalization of α-smooth muscle actin (red) and PDGFRα (green) in mammary glands of nulliparous and InvD6 mice. Boxes show images at high magnification. Arrow: PDGFRα+ cells. Arrowhead: αSMA+ cells. White scale bar: 50 μm. Yellow scale bar: 10 μm. (J) Pathway analysis of RNA-Seq data described in D showing immune regulation–related pathways that are upregulated in InvD6 fibroblasts. (K) RNA-Seq data show chemokine expression pattern of fibroblasts isolated from nulliparous mammary glands. (L) RT-qPCR gene expression for Cxcl12 in sorted PDGFRα+ fibroblasts from nulliparous and InvD6 mammary glands, n = 3–5 per group. Gene expression data are normalized to Actb and are from 2 to 6 independent studies. RNA-Seq data are archived at the NCBI Gene Expression Omnibus (GEO GSE94761). *P < 0.5, **P < 0.01, ***P < 0.001, #P < 0.0001 by unpaired 2-tailed t test. Data represent mean ± SEM.
To determine if mammary fibroblasts from nulliparous mice differed phenotypically from those isolated from actively involuting mammary glands, we performed global gene expression analyses by RNA-Seq on FACS-isolated PDGFRα+ fibroblasts from nulliparous and involution day 6 (InvD6-fibroblasts) mammary glands. We selected InvD6-fibroblasts because the ECM remodeling phase of involution, a putative fibroblast function, peaks at InvD6 (8). Moreover, InvD6 is the peak of immune cell infiltration, and we anticipated that interactions between immune cells and fibroblasts may contribute to fibroblast activation. Over 850 differentially regulated genes were detected between nulliparous- and InvD6-fibroblasts (Figure 1D). Pathway analysis confirmed that ECM protein–associated pathways, especially fibrillar collagen production and ECM remodeling, were upregulated in PDGFRα+ fibroblasts isolated from InvD6 mammary glands compared with mammary fibroblasts isolated from nulliparous hosts (Figure 1E).
To investigate the time course of fibroblast activation during weaning-induced mammary gland involution, we FACS isolated PDGFRα+ mammary fibroblasts from nulliparous and 4-, 6-, and 8-day postweaning (InvD4, InvD6, and InvD8) mice and assessed for fibroblast density and activation markers. The number of PDGFRα+ cells per gram of mammary gland tissue, as determined by FACS analysis, was low in nulliparous mice and at InvD4, but doubled at InvD6 and InvD8 (Figure 1F). Further, on a per-cell basis, Col1a1 and Col3a1 expression levels measured by reverse transcription quantitative PCR (RT-qPCR) were increased across all 3 involution time points compared with nulliparous-fibroblasts, and Col5a1 was increased at InvD4 (Figure 1G). Gene expression for the collagen cross-linking enzyme Lox was highly increased in fibroblasts at InvD4 and InvD6 (Figure 1G), data consistent with elevated fibrillar collagen production. Further, Mmp2, Mmp3, and Tgfb1 expression levels were increased at InvD6 (Figure 1H). These gene expression patterns are consistent with classical fibroblast activation, so we costained mammary tissue for PDGFRα and α-smooth muscle actin (αSMA), a fibroblast activation marker under pathologic conditions (50). αSMA was undetectable in mammary fibroblasts at all reproductive stages (Figure 1I and data not shown).
Another protumorigenic hallmark of mammary gland involution is macrophage and monocyte infiltration, with the peak influx occurring during mid (InvD4 and -6) to late (InvD8 and -10) involution (12, 48). We assessed for immunoregulatory pathways in involution-fibroblasts, and in comparison with nulliparous-fibroblasts, InvD6-fibroblasts had increased gene expression of several immune pathways including rheumatoid arthritis and cytokine-receptor interaction (Figure 1J). A subset analysis of chemokine RNA expression profiles revealed Cxcl12 as the most highly expressed chemokine in these mammary fibroblasts (Figure 1K). CXCL12 is a lymphocyte chemoattractant and immunosuppressive chemokine that induces macrophage M2 polarization and IL-10 production (51–53). Further, CAF-secreted CXCL12 stimulates cancer cell proliferation via paracrine signaling (54). By RT-qPCR, we confirmed that Cxcl12 gene expression was increased in InvD6- compared with nulliparous-fibroblasts (Figure 1L). In summary, the lack of αSMA concurrent with high expression of fibrillar collagens, Lox, Tgfb1, Mmps, and Cxcl12 indicates that involution-fibroblasts are not classically activated, as occurs during wound healing, but rather are alternatively activated through a developmentally regulated process. Further, phenotypic characterization of involution-fibroblasts suggests these cells are capable of modulating 2 key aspects of the tumor microenvironment, fibrillar collagen deposition and the immune milieu.
Ibuprofen inhibits involution-fibroblast activation in vivo.
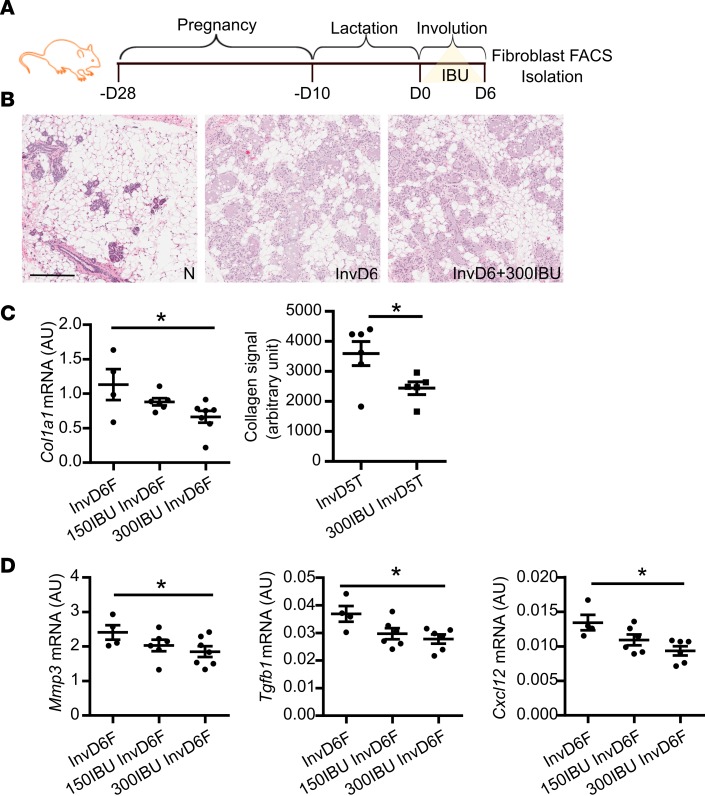
In rodent models, systemic NSAID treatment after weaning inhibits fibrillar collagen accumulation in the involuting mammary gland through an unknown mechanism (5, 55). To determine if systemic NSAID treatment inhibits collagen gene expression in mammary fibroblasts, we treated mice with control diet or ibuprofen at 2 different doses (equivalent to 117 mg and 234 mg per day for humans) (56), starting at weaning through InvD6 (Figure 2A). Based on histologic examination, we found that ibuprofen treatment did not delay normal, postweaning lobule regression (Figure 2B), data consistent with previous studies (5, 6, 32). Ibuprofen treatment also did not influence the number of fibroblasts per gram tissue in the involuting mammary gland (Supplemental Figure 2A). However, assessing FACS-isolated fibroblasts revealed that both doses of ibuprofen decreased fibroblast expression of Col1a1 (Figure 2C), but not Col3a1, Col5a1, or Lox (Supplemental Figure 2B). To determine if this decrease in type I collagen gene expression correlated with decreased protein levels within the mammary gland, type I collagen protein levels were quantified by IHC. This analysis demonstrated decreased type I collagen protein levels after ibuprofen treatment (Figure 2C). Ibuprofen treatment also reduced fibroblast Mmp3 gene expression (Figure 2D), but not Mmp2 expression (Supplemental Figure 2B). Additionally, Tgfb1 and Cxcl12 expression was decreased by ibuprofen treatment (Figure 2D). Cumulatively, these data demonstrate that ibuprofen treatment suppresses key aspects of fibroblast activation including type I collagen production and Mmp2, Tgfb1, and Cxcl12 gene expression.
Figure 2. Systemic in vivo ibuprofen treatment during weaning-induced mammary gland involution decreases mammary fibroblast activation.
(A) Schematic shows breeding and weaning-induced mammary gland involution timeline with 0 mg/kg (control), 150 mg/kg (150IBU), and 300 mg/kg (300IBU) ibuprofen provided in chow for 6 days following weaning, with mammary fibroblasts FACS-isolated on involution day 6 (InvD6). (B) Representative H&E images of nulliparous, InvD6, and 300IBU mammary glands depicting normal nulliparous and InvD6 gland morphology in the absence and presence of ibuprofen treatment. Scale bar: 200 μm. (C) Left: RT-qPCR of Col1a1 gene expression in isolated fibroblasts, n = 4–7 per group. Right: type I collagen IHC quantification around InvD5 mammary lobules (InvD5T) with and without 300IBU treatment for 5 days, n = 5–6 per group. (D) RT-qPCR analysis of targeted genes in isolated fibroblasts, n = 4–7 per group. Gene expression data are normalized to Actb. Ibuprofen intervention data are from 2 independent breeding studies. *P < 0.05 by 1-way ANOVA with Tukey correction and unpaired 2-tailed t test. Data represent mean ± SEM.
Ibuprofen inhibits fibroblast activation in vitro.
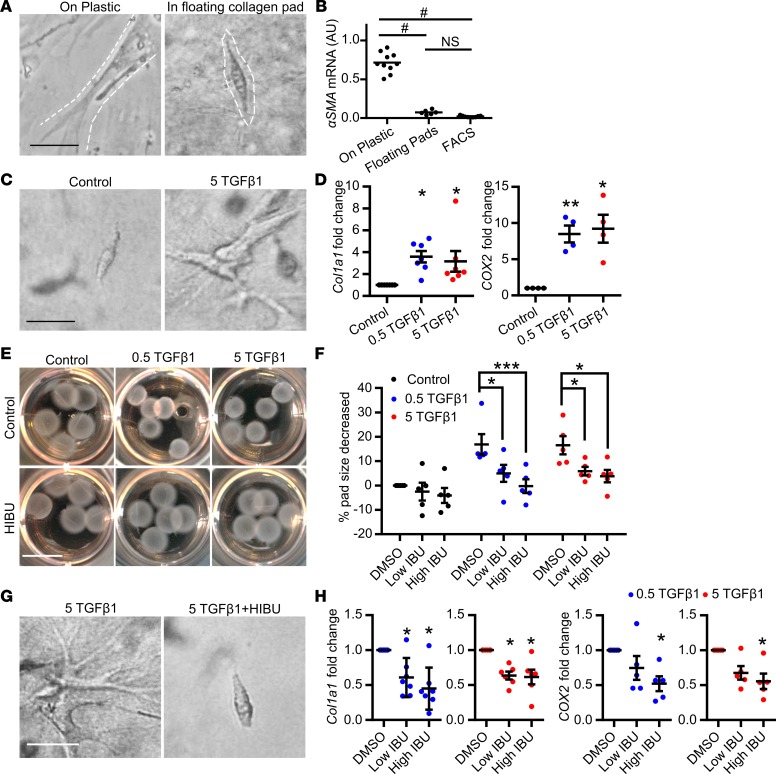
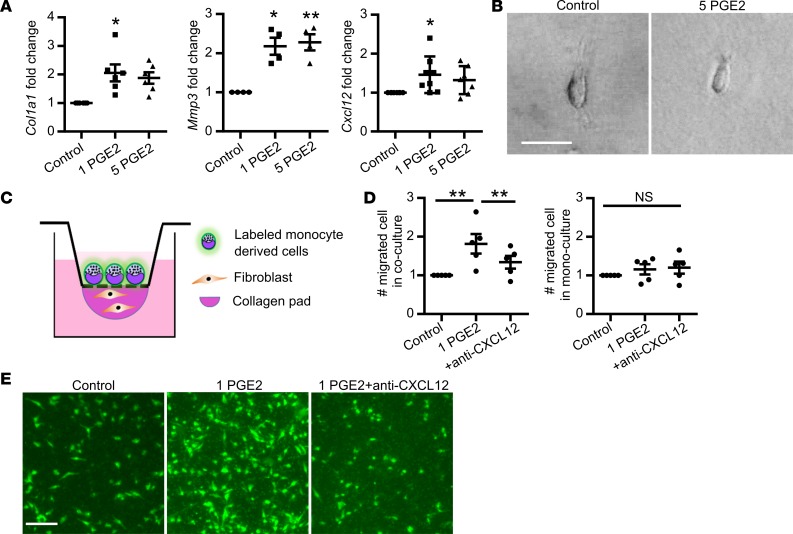
While the above studies demonstrate that systemic ibuprofen treatment reduces mammary fibroblast activation in vivo, these studies do not address whether ibuprofen directly suppresses fibroblast collagen production. To address this question, we isolated mammary fibroblasts from nulliparous mice (57), expanded them in culture, and evaluated their function in an adapted fibroblast contraction model whereby fibroblasts are cultured within a floating collagen pad (58). In comparison with mammary fibroblasts cultured on tissue culture plastic, fibroblasts in the floating collagen pads had a compacted phenotype (Figure 3A) and expressed a low level of αSMA, similar to the level observed in freshly FACS-isolated mammary fibroblasts (Figure 3B). We next modeled the involution microenvironment by treating the fibroblast-impregnated floating collagen pads with TGF-β1, a key mediator of mammary gland involution (59, 60) with established roles in fibroblast activation (61–63). TGF-β1–treated fibroblasts had an elongated phenotype (Figure 3C), increased Col1a1 and COX2 gene expression (Figure 3D), and contracted the collagen pads in a TGF-β1 dose-dependent manner (Figure 3E). Importantly, under these culture conditions, TGF-β1 treatment did not increase fibroblast αSMA expression (Supplemental Figure 3), modeling numerous aspects of the activated fibroblast phenotype observed in the involuting gland. Ibuprofen, added to the cultures at levels detected in the plasma of humans with regular ibuprofen intake (64), inhibited the ability of fibroblasts to contract the collagen pad (Figure 3E and quantification in Figure 3F), elongate (Figure 3G), or induce Col1a1 and COX2 expression (Figure 3H). The ability of NSAIDs to inhibit these fibroblast activation phenotypes suggests that activation is partially dependent on COX2 pathway metabolites. Thus, we treated fibroblasts with the COX2 enzymatic product prostaglandin E2 (PGE2), which is abundant in the involuting mammary gland (32), and found that PGE2 induced fibroblast Col1a1, MMP3, and Cxcl12 expression, but not cellular elongation or pad contraction (Figure 4, A and B).
Figure 3. In vitro mammary fibroblast activation by TGF-β1 is directly inhibited by ibuprofen.
Treatment conditions are: 0.5 TGF-β1 = 0.5 ng/ml TGF-β1, 5 TGF-β1 = 5 ng/ml TGF-β1, Low IBU =10 μg/ml ibuprofen, High IBU = 30 μg/ml ibuprofen (HIBU). (A) Mammary fibroblast morphology when cultured on plastic (left panel) or within floating collagen pads (right panel). Dashed lines show the outline of the cells. (B) α-Smooth muscle actin (αSMA) gene expression in primary mammary fibroblasts cultured on plastic, within floating collagen pads, or from freshly sorted mammary PDGFRα+ fibroblasts, n = 6–10 per condition. (C) Morphologic evidence of TGF-β1–induced fibroblast activation in the floating collagen pad culture model. (D) Increased Col1a1 and Cox2 gene expression in fibroblasts treated with TGF-β1, n = 4–7 per condition. (E) The ability of TGF-β1–treated fibroblasts to contract collagen pads (top) is suppressed by ibuprofen (bottom) and (F) data quantification, n = 5 per condition. (G) Fibroblast morphology with TGF-β1 treatment in the presence or absence of ibuprofen. (H) Col1a1 and Cox2 gene expression in fibroblasts with TGF-β1 treatment in the presence or absence of ibuprofen, n = 5 per condition. Scale bars: 100 μm (A, C, and G) and 1 cm (E). All gene expression data are normalized to Actb and then normalized to the control groups in each experiment. TGF-β1 and ibuprofen combination treatment studies were repeated 5 times. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001 by 1-way ANOVA with Tukey correction. For data normalized to control, statistics were performed using matched 1-way ANOVA with Tukey correction on the raw data that are not normalized to control. Data represent mean ± SEM. NS, not significant.
Figure 4. Prostaglandin E2 (PGE2) directly stimulates fibroblast collagen and cytokine expression but not cell elongation.
(A) Col1a1, Mmp3, and Cxcl12 gene expression induced by 1 ng/ml PGE2 (1 PGE2) or 5 ng/ml PGE2 (5 PGE2) treatment, n = 4–8 per group. All gene expression data are normalized to Actb and then normalized to the control groups in each experiment. Gene expression data obtained from 4 to 8 independent experiments. (B) Mammary fibroblast morphology when cultured within floating collagen pads in the absence (right) and presence of 5 ng/ml PGE2. Scale bar: 100 μm. (C) Model of fibroblast-induced monocyte-derived cell migration assay setup. (D) Quantification of the number of monocyte-derived cells that migrated into collagen pads in Control (1 μg/ml mouse IgG1 isotype control), 1 PGE2 (1 ng/ml PGE2 with 1 μg/ml mouse IgG1 isotype control), and +anti-CXCL12 (1 ng/ml PGE2 with 1 μg/ml CXCL12-neutralizing antibody) conditions, with data normalized to control, n = 5/condition. Monocyte-derived cell migration assay was repeated 5 times. (E) Representative images of migrated monocyte-derived cells labeled with CFSE (green) fluorescent dye in Control, 1 PGE2, and 1 PGE2+anti-CXCL12 conditions as described in D. Scale bar: 200 μm. *P < 0.5, **P < 0.01 by matched 1-way ANOVA with Tukey correction on the raw data that are not normalized to control. Data represent mean ± SEM. NS, not significant.
Fibroblasts recruit monocytes in a COX/PGE2–dependent manner in vitro.
The induction of Cxcl12 in PGE2-treated fibroblasts (Figure 4A) supports the hypothesis that PGE2-activated fibroblasts may mediate chemotaxis of monocyte-derived cells, an immune cell abundantly present in the involuting mammary gland (7). To begin to test this hypothesis, we utilized a modified cell migration assay, in which collagen pads containing fibroblasts were separated from fluorescently labeled bone marrow–derived monocytes by a membrane with 5-μm pores. Fibroblasts were then treated with vehicle (control), PGE2, or PGE2 with CXCL12-neutralizing antibody, and the impact on monocyte-derived cell recruitment into the collagen pad was quantified (Figure 4C). We found that PGE2 treatment of fibroblasts induced monocyte-derived cell migration, and further, found that the addition of CXCL12-neutralizing antibody completely reversed this phenotype (Figure 4, D and E). In the absence of fibroblasts, monocyte-derived cells failed to migrate in response to PGE2, demonstrating the requirement of fibroblasts for monocyte-derived cell migration in this assay (Figure 4, D and E). Collectively, our cell culture studies demonstrate a role for PGE2 in fibroblast activation and subsequent monocyte-derived cell recruitment via CXCL12.
Ibuprofen inhibits tumor promotional capacity of involution-fibroblasts.
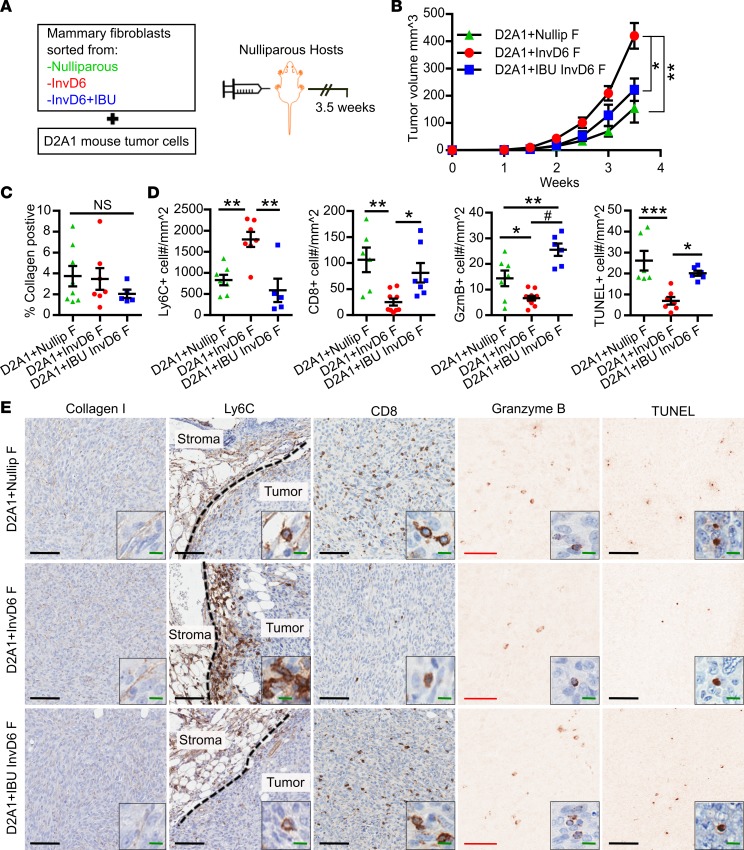
To determine the functional significance of mammary fibroblast activation state on mammary cancer promotion, we orthotopically injected BALB/c mice with D2A1 mammary tumor cells mixed 1:1 with FACS-isolated mammary fibroblasts from mice that were nulliparous, InvD6, or InvD6 treated with ibuprofen at weaning through InvD6 (IBU-InvD6-fibroblasts) (Figure 5A). Sorted fibroblasts, plated in culture, confirmed uniform cell viability across groups (Supplemental Figure 4). Compared with nulliparous-fibroblasts, D2A1 mammary tumor cells coinjected with InvD6-fibroblasts formed larger tumors (Figure 5B). Tumors that developed from coinjection with IBU-InvD6-fibroblasts were smaller, similar in size to the nulliparous-fibroblast group tumors (Figure 5B). At study end we found no significant difference in intratumoral collagen deposition between nulliparous- and InvD6-fibroblast group tumors (Figure 5, C and E). However, we did find evidence supportive of involution-fibroblasts mediating a protumorigenic, immunosuppressive milieu, as tumors in the InvD6-fibroblast group had increased Ly6C+Ly6G– monocyte density at the tumor border (Figure 5, D and E, and Supplemental Figure 5A), and decreased intratumoral CD8+ T cell and granzyme A+ and granzyme B+ cell infiltrates (Figure 5, D and E, and Supplemental Figure 5, D and E). In contrast, tumors in the IBU-InvD6-fibroblast group had decreased Ly6C+ monocytes at the tumor border, increased intratumoral CD8+ T cells, and increased granzyme A+ and granzyme B+ cell infiltrates (Figure 5, D and E, and Supplemental Figure 5, D and E). Costaining of CD8 and granzyme B demonstrated a subset of granzyme B+ CD8+ T cells, consistent with the presence of activated, intratumoral CD8+ T cells in the IBU-InvD6-fibroblast group tumors (Supplemental Figure 5, B and C). However, the identity of the immune cells producing granzyme A remains unknown. By IHC we found these granzyme A+ cells to be CD45+CD3–CD8–CD4–NCR1–, suggesting they are not T cells, NKT cells, or NK cells (Supplemental Figure 5F). No differences in Foxp3+ T regulatory cells were observed between groups (Supplemental Figure 6, A and B). Further, in spite of significant tumor size variation between groups, we did not observe differences in tumor cell proliferation (Supplemental Figure 6C). However, we did observe differences in tumor cell death, with InvD6-fibroblasts associated with decreased death and IBU-InvD6-fibroblasts with increased death (Figure 5, C and D). Cumulatively, our combined studies suggest fibroblast activation during mammary gland involution that is mediated through a COX-dependent mechanism. These activated fibroblasts recruit Ly6C+ monocytes via CXCL12 secretion, which in the presence of tumor, inhibit CD8+ T cell tumor infiltration and tumor cell death, resulting in enhanced tumor size (Figure 6).
Figure 5. Mammary involution-fibroblasts are tumor promotional via a COX2-dependent mechanism that suggests immune modulation.
(A) Schematic design for mammary fibroblast treatment, isolation, and use in an orthotopic mammary tumor model. FACS-isolated mammary fibroblasts (20,000 cells) from nulliparous, involution day 6 (InvD6), or InvD6 hosts treated during involution with 300 mg/kg ibuprofen in chow (IBU InvD6 F), were mixed with D2A1 tumor cells at a 1:1 ratio, injected into mammary fat pads of nulliparous BALB/c mice, and tumor tissue collected 3.5 weeks after tumor cell injection. (B) Tumor growth curve of D2A1 cells coinjected with sorted fibroblasts from nulliparous (D2A1+Nullip F), InvD6 (D2A1+InvD6 F) and InvD6 with ibuprofen treatment (D2A1+IBU InvD6 F) mice, n = 7–10 tumors per group. (C) IHC quantification of intratumoral type I collagen, n = 5–8 tumors per group. (D) IHC quantification of tumor border Ly6C, intratumoral CD8, granzyme B (Gzmb), and TUNEL; n = 5–10 tumors per group. (E) Representative IHC images of markers quantified in D. From left to right: intratumoral collagen I, tumor border Ly6C (dashed lines show tumor border), intratumoral CD8, granzyme B, and TUNEL. Black scale bars: 100 μm. Red scale bars: 50 μm. Green scale bar: 10 μm. Counterstaining (blue) is not shown in granzyme B– and TUNEL-stained images to enhance visualization of the positive cells. All tumor data were obtained from 2 independent breeding studies. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001 by 1-way ANOVA with Tukey correction. Data represent mean ± SEM. NS, not significant.
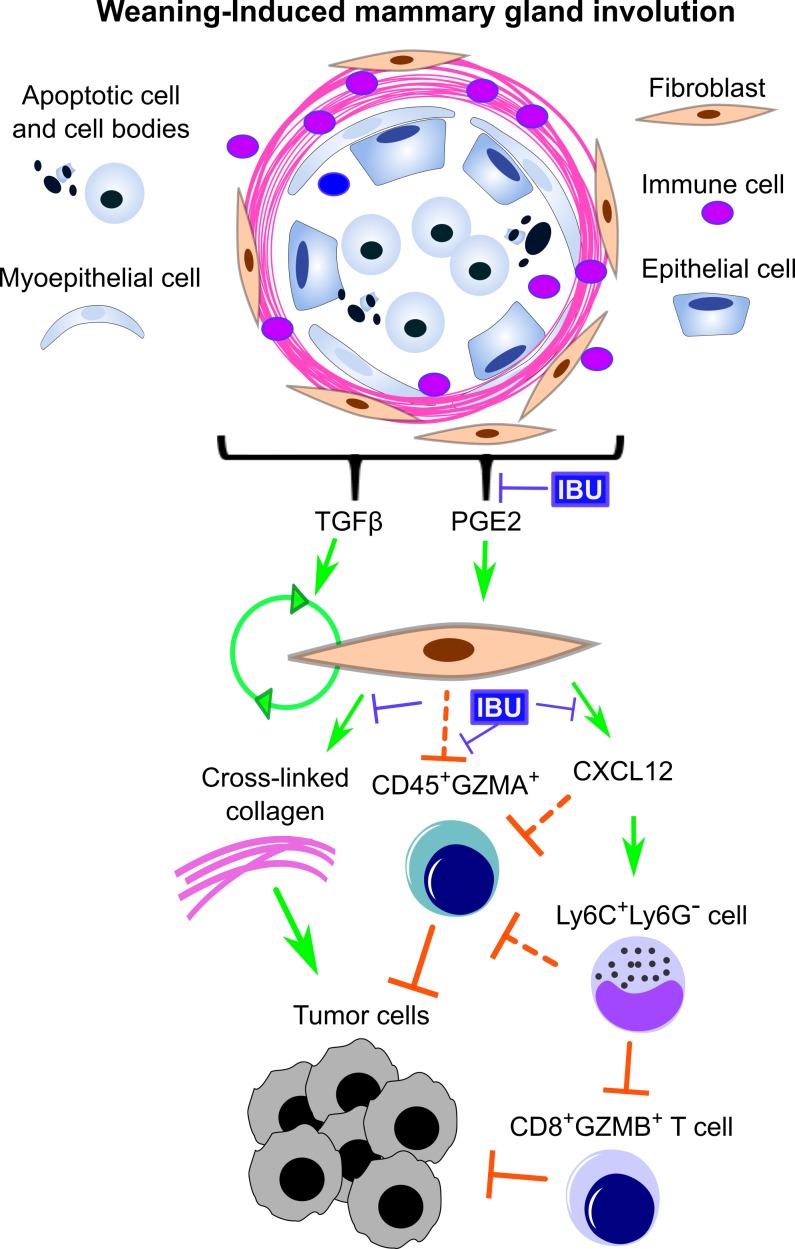
Figure 6. Schematic overview of the potential tumor promotional contributions of mammary involution-fibroblasts.
Activation of involution-fibroblasts by TGF-β and prostaglandin E2 (PGE2) in the involution microenvironment leads to increased fibrillar collagen expression, which is tumor promotional. Involution-fibroblasts also have increased CXCL12 expression, which recruits Ly6C+Ly6G– monocytes, blocking CD8+ T cell tumor infiltration and tumor cell death. Involution-fibroblasts also block CD45+GZMA+ cell infiltration into tumors by an unknown mechanism (dashed block arrows). These tumor promotional attributes of the involution-fibroblasts can be blocked by ibuprofen (IBU). GZMA and GZMB, granzymes A and B.
Discussion
We identify, potentially for the first time, a developmentally activated PDGFRα+ fibroblast population in the postweaning rodent mammary gland that shares numerous attributes with myofibroblasts activated under wound healing and cancer pathologies. These PDGFRα+ fibroblasts are characterized by increased gene expression for the fibrillar collagens Col1a1, Col3a1, and Col5a1, and the stromal remodeling factors Tgfb1, Lox, Mmp2, Mmp3, and Cxcl12. Unlike fibroblasts activated under pathological conditions, mammary involution-fibroblasts have low αSMA expression, suggesting a previously unrecognized activation state. Further, we find involution-fibroblasts to be tumor promotional, as they increase mammary tumor growth in an immunocompetent, orthotopic tumor transplant model. These rodent studies are consistent with the concept that breast cancers can misappropriate normal developmentally regulated programs to their advantage, such as physiologic fibroblast activation during mammary gland involution. Young women’s breast cancer may be especially responsive to such developmentally regulated programs owing to the remarkable, physiologic breast remodeling that accompanies gland regression in the postpartum setting (12, 13).
The links between NSAIDs, COX2, mammary gland collagen density, and breast cancer progression were first reported in a model of postpartum breast cancer (5). More recently in the Col1a1tm1jae mouse model of high breast density, where a mutation in the collagen I collagenase cleavage site leads to increased collagen accumulation and subsequent mammary tumor promotion, celecoxib was found to suppress both phenotypes (55). Here, possibly for the first time, we identify a potential mechanism by which NSAIDs inhibit collagen deposition by demonstrating that the COX2 metabolite PGE2 induces, and ibuprofen suppresses, Col1a1, Mmp3, and Cxcl12 production in normal mammary fibroblasts. Given the paucity of data pertaining to physiologic collagen regulation in the breast, we propose that weaning-induced mammary gland involution may be a useful model to study the role of fibroblast activation and collagen production under normal physiological conditions. Such studies may provide insight into the regulation of breast density, a dominant risk factor for breast cancer (65), as well as breast cancer progression (27).
One unanswered question pertains to the source of COX2 metabolites in the involuting mammary gland. Surprisingly, mammary epithelial cells (MECs) express high levels of COX2 during weaning-induced involution and MECs may be the dominant source of PGE2 in the mammary gland (66). Supportive roles for MECs in fibroblast activation and collagen production have been previously reported. For example, overexpression of p190B RhoGAP (GTPase activating protein) in MECs has been shown to increase TGF-β secretion and subsequent fibroblast activation (67). Additionally, MEC-derived pappalysin-1, which is upregulated during weaning-induced involution, releases bound IGF through cleavage of the IGF binding proteins 4 and 5. The resulting elevated IGF signaling stimulates collagen production in mammary fibroblasts (68, 69). It remains to be determined whether the COX2/PGE2 pathway we describe here for mammary fibroblast activation integrates with either the p190B RhoGAP or pappalysin-1 pathways during weaning-induced involution.
Having demonstrated that involution-fibroblasts upregulate fibrillar collagen expression, it was surprising that there were no observable increases in collagen abundance in mammary tumors that developed after coinjection with InvD6-fibroblasts. One limitation of our tumor study is that we did not determine the fate of the injected primary fibroblasts over time, which are nontransformed and likely did not survive to study end. Early loss of fibroblasts in our transplant model may explain the unchanged collagen levels across groups. However, tumor sizes did differ between fibroblast groups, raising the question of whether early collagen exposure contributes to differences in tumor size across groups. One possible explanation, consistent with an early role for collagen in tumor expansion, is that transient exposure to fibroblast collagen durably influences tumor cell behavior. In support of this possibility, in an orthotropic model, compared with ECM isolated from nulliparous mammary glands, collagen-rich ECM isolated from involuting mammary glands promoted mammary tumor development for several weeks, even at ECM volumes that limited direct tumor cell–ECM interactions to only a few days after tumor cell injection (32, 70). In further support of a durable protumor effect resulting from transient exposure to the involuting microenvironment, mammary tumor cells that evolved for 2 weeks within the involuting gland retained elevated expression of COX2 and prometastatic attributes ex vivo (6). Of potential relevance, durable, protumorigenic effects have been reported after transient activation of the Src oncoprotein in immortalized breast MCF10A cells (71). In this report, stable cellular transformation was maintained in the absence of the initiating oncogenic signal due to establishment of a positive feedback loop involving NF-κB, IL-6, and STAT3 (71). Such feed-forward loops may explain, in part, how fibroblast collagen gene expression at the time of tumor cell injection might impact tumor size, even in the absence of differences in tumor collagen abundance at study end. However, additional studies are required to test this hypothesis.
We also found that mammary fibroblasts isolated from involuting mammary glands had immunomodulatory function consistent with immune suppression. Immunomodulatory activity of CAFs has been previously reported (21, 23), and shown to be mediated in part by cytokine and chemokine secretion. Additional indirect evidence for immunoregulatory functions of CAFs is suggested by the positive correlations between the high ECM deposition by CAFs, tumor immune cell infiltration, and breast cancer progression in women (72, 73). Here, we show for possibly the first time that physiologically activated fibroblasts, weaning-induced involution-fibroblasts, characterized by high ECM production and chemokine secretion, also have immunomodulatory functions. Previous studies of the involuting mammary microenvironment have identified increased M2-polarized macrophages (arginase 1 positive), immature monocytes that suppress T cell CD25, CD69, and IFN-γ production, and increased levels of immunosuppressive cytokines, including IL-4, IL-13, and IL-10 (7, 11, 12, 74). Here, we extend these studies to show that the chemokine CXCL12, a powerful monocyte lineage chemoattractant (51), is produced by fibroblasts during mammary involution, potentially in response to increased PGE2 signaling, as in vitro, PGE2 treatment induced fibroblast Cxcl12 expression and monocyte-derived cell recruitment. Further, we show that fibroblast Cxcl12 expression levels correlate with the presence of tumor-associated monocytes at the tumor border, and inversely with tumor-infiltrating CD8+ T cells, 2 critical immune cell populations controlling tumor cell death (75). How fibroblasts influence CD8+ T cell infiltration and tumor cell death in this transplant model of postpartum breast cancer remains to be determined.
One limitation of our study is that by using PDGFRα positively selected mammary fibroblasts, we may be evaluating a subset of mammary fibroblasts, as other PDGFRα– fibroblasts have been reported, such as in bone marrow (76). Further, while we confirmed these PDGFRα+ cells to be largely devoid of epithelial, immune, and endothelial cell type markers, this population likely includes pericytes and perivascular fibroblasts (77, 78). Another limitation is that we restricted our functional analyses to nulliparous and InvD6 mammary fibroblasts, which does not fully capture the dynamic nature of fibroblasts in the involution window, nor the duration of fibroblast activation. Further, we have not evaluated the phenotype of mammary fibroblasts during pregnancy, another dynamic tissue remodeling window, due to our focus on postpartum breast cancer (1,2).
With respect to postpartum breast cancer in women, an unanswered question is how the transient event of weaning-induced involution contributes to the poor prognosis of breast cancers diagnosed a full 5–10 years after childbirth. As discussed above, one possible mechanism is the establishment of positive feed-forward loops that persist in the primary tumor microenvironment after completion of postpartum breast involution. An additional unexplored mechanism is that the involution microenvironment stimulates existing indolent tumor cells to disseminate at early stages. The indolent primary tumor and the disseminated tumor cells might then require additional years of evolution to become clinically detectable. Postpartum models that encompass clinically relevant early stage breast disease are required to address these important questions.
Our data suggest the fibroblast as a potential therapeutic target for postpartum breast cancer patients. However, the balance of fibroblast function needs to be carefully considered as it is becoming increasingly clear that fibroblasts exhibit a range of activities from tumor promotional to tumor suppressive, similar to that observed for immune cells (79). On one side of this functional spectrum, numerous robust studies report the ability of pathologically activated fibroblasts to support cancer progression (80–82). Further, our observation that normal mammary fibroblasts can be tumor promotional, combined with published data showing that senescent fibroblasts associated with natural aging promote breast cancer (83), demonstrate how physiologic states might contribute to protumorigenic fibroblasts. In sum, these protumorigenic fibroblast studies suggest that fibroblast ablation in the tumor microenvironment might have therapeutic merit. However, a recent, paradigm-shifting study showed that depletion of murine pancreatic cancer fibroblasts increased tumor metastasis and decreased survival (84). Thus, therapies directed at fibroblasts need to be focused on modulating the balance towards tumor-suppressive activity rather than ablation, as recently proposed (21).
In summary, we describe a physiologically activated fibroblast with tumor promotional attributes that is present within the involuting mammary gland following weaning. Systemic treatment with ibuprofen after weaning reverts this activated and tumor promotional phenotype to baseline, nulliparous levels, identifying fibroblast activation as a target of NSAIDs, possibly for the first time. Further, our finding of fibroblast NSAID responsivity raises the interesting hypothesis that the ability of NSAIDs to inhibit breast cancer (85, 86) may comprise targeting the mammary stroma, including fibroblast-mediated collagen deposition and immune suppression. Finally, postpartum breast cancer may be uniquely driven by a collagen-rich and immune-suppressed microenvironment (5, 7, 12), and thus particularly responsive to NSAID intervention; identifying NSAIDs as a treatment strategy warrants further investigation.
Methods
Mouse breeding, reproductive staging, and histology.
Eight-week-old female BALB/c mice (Jackson Laboratory) were housed, bred, and pup number normalized as described (12), with age-matched littermate nulliparous mice for controls. To initiate involution, pups were weaned between 9 and 13 days of age. Mice were euthanized on designated day of involution and left and right inguinal mammary glands harvested. For histologically based assays, the region of the gland with associated lymph nodes was dissected, fixed in 10% neutral buffered formalin (Anatech Ltd.) for 48 hours and then processed, paraffin embedded, and sectioned at 4 μm. The confirmation of normal histology was performed by H&E staining. The lymph node–free mammary tissue was either stored in cold Hank’s Balanced Salt Solution for flow analysis or snap-frozen in liquid nitrogen and stored at –80°C for RNA and protein analyses. For mice receiving ibuprofen, treatment was initiated at the day of weaning and continued for 6 days of treatment, at which time tissues were harvested. Ibuprofen (Sigma-Aldrich, I7905) was delivered at 150 mg/kg or 300 mg/kg of chow with mice fed ad libitum.
Tissue processing, FACS, and tumor model.
Mammary glands from BALB/c mice were harvested and processed as previously described (46). PDGFRα antibody (BioLegend, 135910) was used at 1:25 to stain fibroblasts for FACS. The purity of isolated fibroblasts was greater than 90% as evaluated by flow cytometry using PDGFRα. Sorted fibroblasts were either frozen at –80°C for gene expression analysis or mixed with 20,000 D2A1 cells at a 1:1 ratio in 10 μl PBS and injected into the left and right fourth mammary glands of BALB/c nulliparous mice (12 weeks old), under isoflurane (Terrell) anesthesia. Tumor length (L) and width (W) were measured twice per week using an electronic caliper. Tumor size was calculated as (L × W2)/2. Mice were sacrificed at 3.5 weeks after injection for tissue collection with tumors fixed in 10% neutral buffered formalin for 48 hours and then processed, paraffin embedded, and sectioned at 4 μm for staining.
RNA isolation and RT-qPCR analysis.
RNA was isolated from sorted fibroblasts (Macherey-Nagel, 740955), or floating collagen pads (Invitrogen, 10296-028; QIAshredder, 79656; Invitrogen, 12183-016). To ensure adequate RNA yield, more than 0.1 million FACS-isolated PDGFRα+ cells were used for RNA isolation and hence mammary glands were pooled from 5 or 6 nulliparous mice and 2 or 3 involution mice (InvD4, -6, and -8). RNA concentration and purity were determined by absorbance at 260 nm, 230 nm, and 280 nm. RNA quality was determined by an Agilent 2100 Bioanalyzer. cDNA was synthesized using a Bio-Rad cDNA synthesis kit and qPCR was performed using Bio-Rad SYBR Green supermix with MyiQ Single Color Real-Time PCR Detection System. Mouse primer sequences were: reference gene β-actin (Actb), forward GCAACGAGCGGTTCCG and reverse CCCAAGAAGGAAGGCTGGA; Col1a1, forward TTCTCCTGGCAAAGACGGACT and reverse AGGAAGCTGAAGTCATAACCG; Col3a1, forward AGCTTTGTGCAAAGTGGAACC and reverse CAAGGTGGCTGCATCCCAATT; Col5a1, forward TCTCTGTGTGTGTGCCAAGAT and reverse AGCCAGAGTCCATCCCACATT; lysyl oxidase, forward CAAGGGACATCGGACTTCTTAC and reverse TGGCATCAAGCAGGTCATAG; αSMA (Acta1), forward CCATCATGCGTCTGGACTT and reverse GGCAGTAGTCACGAAGGAATAG;Tgfb1, forward TACGTCAGACATTCGGGAAGCAGT and reverse AAAGACAGCCACTCAGGCGTATCA; Mmp2, forward AGTGGTCCGCGTAAAGTATGGGAA and reverse ATTGCCACCCATGGTAAACAAGGC; Mmp3, forward TGGAACAGTCTTGGCTCATGCCTA and reverse TGGGTACATCAGAGCTTCAGCCTT; COX2, forward GCATTCTTTGCCCAGCACTTCACT and reverse TTTAAGTCCACTCCATGGCCCAGT. All primers were purchased from Integrated DNA Technologies. RT-qPCR conditions were as follows (except for COX2): 95°C for 3 minutes, then 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. For COX2 qPCR, QuantiTect SYBR Green (Qiagen, 204243) was used. The conditions for COX2 qPCR were 95°C for 15 minutes, then 40 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 30 seconds.
RNA-Seq analysis.
Total RNA was purified as described above. mRNA isolation and library preparation were performed by the Massively Parallel Sequencing Shared Resource at Oregon Health & Science University, using a TruSeq RNA Sample Preparation V2 kit. Four samples were pooled per lane and sequenced on an Illumina HiSeq 2500 with 100-bp single-ended reads. The resulting data were converted to FASTQ format using Bcl2Fastq software (Illumina). Reads were aligned to the mouse reference genome, build GRCm38, using STAR 2.4.2a. STAR performed counting of reads per gene as defined in Ensembl build 81 (GENCODE version m6). Read counts were adjusted to a common non-zero minimum via addition of uniform minor background counts. Read count distributions were normalized across samples using cyclic Loess normalization by R software. Normalized read counts were log2-transformed for further analysis. Differential expression was determined by fitting linear regression models for each log2-normalized gene expression level with InvD6 as the independent variable. A Wald test was performed to test the null hypothesis of no association of involution with expression for each gene. Multiple hypothesis testing was accounted for by controlling the false discovery rate (FDR) at 5% with the q-value approach (87). The final set of significant genes was further filtered to include only genes with absolute value of log2-fold change greater than 0.4 (1.3-fold). Power analyses were performed via simulations based on the variability of expression and magnitude of fold changes observed in the current data. The analysis methods were assumed to be identical to the methods above. Power analyses were performed under the alternative hypothesis of 500 truly differentially expressed genes with absolute value log2-fold changes greater than 0.7. Unsupervised clustering was performed on the set of differentially expressed genes to generate a heatmap of normalized expression values, centered and scaled within each gene. Pathway analyses were also performed, and the hypergeometric test was performed with Holm adjustment for multiple testing to control the family-wise error rate within an annotation source. This commonly used competitive test examines the odds of finding at least the observed number of significant genes in the pathway while the remainders not in the pathway are greater than expected by chance. The upper tail of the hypergeometric test was used to determine whether the number of selected genes also present in a pathway-based gene set is greater than expected by chance. The sets of pathway annotation sources queried were KEGG, The Broad Institute’s Molecular Signatures Database v5, Gene Ontology Biological Process, Gene Ontology Molecular Function, Gene Ontology Cellular Component, Pathway Commons, BioCarta, and Reactome. Analyses and annotations were performed using custom R scripts and Bioconductorthe R packages “data.table”, “AnnotationDbi”, “XML”, and “GOdb”.
Immunofluorescence.
Formalin-fixed tissue was paraffin embedded (FFPE) and sectioned at 4 μm. Slides were pretreated in Target Retrieval Solution, pH 9 (Dako, S2367) at 125˚C under pressure for 5 minutes. Tissues were blocked with 0.6% H2O2 (diluted in PBS) for 10 minutes, and then treated with protein block (Dako, X0909) for 10 minutes. The staining was performed using Opal 3-Plex kit (PerkinElmer). Primary antibodies used were: anti-PDGFRα (R&D Systems, AF1062) diluted 1:50 for 2 hours at room temperature; anti-GFP (Vector, BA0702) diluted 1:1,000 for 1 hour at room temperature; anti-calponin (Abcam, ab46794) diluted 1:600 for 16 hours at 4˚C; and anti-αSMA (Dako, M0851) diluted 1:400 for 30 minutes at room temperature. Secondary antibodies used were: anti-goat (Santa Cruz Biotechnology, SC-2922) diluted 1:100; streptavidin-HRP (DAKO, P0397); anti-rabbit (Thermo Fisher Scientific, 31461) diluted 1:500; and anti-mouse (Dako, K4001). All secondary antibody incubations were for 1 hour at room temperature.
Immunohistochemistry.
Sections (4 μm) of FFPE mouse mammary glands were used for IHC. Pretreatment: For collagen I, slides were pretreated with Citra buffer (BioGenex, HK086) in a microwave at 1,100 W for 2 minutes. For Ly6C, CD8, and granzyme B, slides were pretreated with Target Retrieval Solution (Dako, S1699) at 125˚C under pressure for 5 minutes. Pretreated slides were then treated with 0.6% H2O2 in methanol (for CD8) or 3% H2O2 in methanol (for all other targets) for 10 minutes and then blocked with protein block (Dako, X0909) for 1 hour. Primary and secondary antibodies used were: anti–collagen I (Abcam, ab34710, 1:100), followed by DAKO EnVision+ Peroxidase Rabbit (DAKO, K4003); anti-Ly6C (Abcam, ab15627, 1:1,500), followed by Rat-on-Mouse HRP-Polymer (Biocare, RT517); anti-CD8 (eBioscience, 14-0808-82, 1:50), followed by Histofine Simple Stain anti-Rat; and anti–granzyme B (Abcam, ab4059, 1:300) followed by Histofine Simple Stain anti-Rabbit. All primary antibodies were incubated for 1 hour and secondary antibodies were incubated for 30 minutes, followed by incubation with the chromogen 3,3′-diaminobenzidine (Dako, K346811) for 10 minutes and counterstained in hematoxylin for 6 minutes. For TUNEL staining, the TACS 2 TdT-DAB In Situ Apoptosis Detection Kit (from Trevigen) was used with Mn2+ cation at a final concentration of 1× as per the manufacturer’s instructions. Protocol and antibody information for data in supplemental figures are in the supplemental methods section. After staining, slides were scanned with an Aperio ScanScope AT, and IHC signal quantification performed by Aperio ImageScope v12.1.0.5029 as described previously (5). The tumor border was defined as 100 μm from the tumor edge for the Ly6C quantification study.
Cell culture.
The mouse tumor cell line D2A1, a gift from Ann Chambers (London Regional Cancer Program, London Health Sciences Centre, London, UK), was obtained from Jeffrey E. Green (Laboratory of Cell Biology and Genetics, National Cancer Institute, Bethesda, Maryland, USA). D2A1 cells were cultured on tissue culture plastic in DMEM medium (HyClone, SH30022.01) supplemented with 10% FBS (HyClone, SH30071.03) and 1× penicillin-streptomycin solution (Corning, 30-002-CI). D2A1 cells were cultured for 48 hours before mammary fat pad injection. Primary mouse mammary fibroblasts were acquired by first plating nulliparous mouse mammary gland digest (2.5 mg/ml collagenase II and IV Worthington, 37°C for 30 minutes) on tissue culture plastic or on polydimethylsiloxane-coated (SYLGARD, 184 Silicone Elastomer Kit) plastic, and supplied with DMEF-12 medium (HyClone, SH30023.01) and 10% FBS (57). Under long-term culture, after 9 days, fibroblasts were highly enriched. These fibroblasts were passaged 3 times to expand the population before freezing in liquid nitrogen. For culturing fibroblasts in floating collagen pads, frozen fibroblasts were thawed and plated on polydimethylsiloxane-coated plates supplied with DMEF-12 medium and 10% FBS for 48 hours, and then changed to DMEF-12 medium with 1% FBS and 200 μM ascorbic acid (Sigma-Aldrich, A0278) and 1× penicillin-streptomycin (fibroblast base medium) for 4 to 6 days until cells reached 80% confluence. Fibroblasts were then mixed with 2 mg/ml collagen I (Corning, 354249) and plated in a 96-well plate at a concentration of 20,000 cells per 50 μl collagen pad. Collagen pads were supplied with fibroblast base medium with the following additives: 0.5 ng/ml or 5 ng/ml TGF-β1 (eBioscience, 14-8342-62), 10 μg/ml or 30 μg/ml ibuprofen (Sigma-Aldrich, I7905), 1 ng/ml or 5 ng/ml PGE2 (Sigma-Aldrich, P6532). After 18 hours in culture, collagen pads were dissociated from the 96-well plate and put in a 12-well plate. After another 30 hours, pictures were taken and the pad diameter measured using an Aperio ImageScope and then 3 to 5 pads of the same condition were pooled for RNA isolation.
Monocyte-derived cells.
Monocyte-derived cells were generated according to previous publications (88, 89). Briefly, bone marrow cells were isolated from femurs and tibias of 10-week-old female BALB/c mice, plated in 6-cm Petri dishes at 10 million cells/plate, and cultured in RPMI medium (HyClone, SH30096.01) supplied with 5% FBS, 1× penicillin/streptomycin, and 30% conditioned medium from the supernatants of M-CSF–secreting L929 fibroblasts (90), a gift from Peter Henson (National Jewish Health). After 48 hours, the plates were washed gently twice with PBS to eliminate dead cells, and then fresh medium with 10 ng/ml mouse recombinant IL-4 (PeproTech, 214-14) was added. After 24 hours, monocyte-derived cells were harvested by gently pipetting to collect only the loosely adhered cells, as previously described (88). Using this protocol and as determined by flow analysis (Supplemental Figure 7) we obtained a mixture of Ly6C+ monocytes (30%), macrophages (25%), and immature precursors (45%) (91). We refer to this cell population as monocyte-derived cells. Detailed methods are provided in the supplemental methods section.
Monocyte-derived cell migration assay.
Fibroblast collagen pad culture was described above. Fibroblasts (1.6 × 104) were mixed in 40 μl of 2 mg/ml collagen and plated on a transwell insert with a pore size of 5 μm (Millipore, MCMP24H48), and then placed inverted in a 12-well plate and allowed to solidify for 1 hour in a 37˚C incubator. The inserts were placed in 24-well plates right-side up, supplied with fibroblast base medium with the addition of 1 ng/ml PGE2, 1 μg/ml CXCL12-neutralizing antibody (R&D Systems, MAB310), and/or 1 μg/ml mouse IgG1 isotype control (R&D Systems, MAB002) and cultured for 24 hours prior to the addition of monocyte-derived cells. The monocyte-derived cells were generated as described above and labeled with CellTrace CFSE (Invitrogen, C34554) at a concentration of 1.25 μM in PBS for 20 minutes at room temperature, and then 7.5 × 104 cells were plated on top of each insert. The medium was changed immediately after plating monocyte-derived cells, using the same medium described above for fibroblasts but with the addition of 1% L929-conditioned medium in all conditions. After 15 hours, the cultures were fixed in Cytofix fixation buffer (BD, 554655) for 20 minutes. The number of fluorescently labeled monocytes that migrated to the bottom of the filter was detected using a Nikon Eclipse TE200 fluorescence microscope. Monocyte-derived cells were first visualized to assure monocyte plating at the same confluence across conditions. After removal of cells on the top of the insert by cotton swab, 4 or 5 pictures at ×4 magnification were taken for each insert to detect migrated cells. Images were processed using ImageJ software (NIH). Briefly, fluorescence intensity was measured by ImageJ and the fluorescence threshold was adjusted by the mean intensity + 2-fold standard deviations. For images with high background, fluorescence threshold was adjusted by mean intensity + 3-fold standard deviations. Cell number was counted by ImageJ “Analyze particles” function. For images with severe background, cell numbers were counted by hand, with data independently verified under conditions where the reader was blinded to study design. The same cell counting methods were used across groups in each experiment.
Statistics.
Unpaired 2-tailed t test and 1-way ANOVA with Tukey correction were performed in GraphPad Prism, assuming independent samples and normal distributions. Only P values less than 0.05 were considered significant. Statistical analysis for the RNA-Seq data set is described above.
Study approval.
Animal procedures were approved with ethical consideration by the University of Colorado Anschutz Medical Campus and Oregon Health & Science University (OHSU) Institutional Animal Care and Use Committees.
Author contributions
Q. Guo and P. Schedin developed the hypothesis, conceptual and experimental designs, and are responsible for data analysis and interpretation. Q. Guo performed all experiments and P. Schedin is responsible for data integrity. For the RNA sequencing study, P. Spellman assisted with experimental design, K. Chiotti processed the raw RNA sequencing data, J. Burchard performed data normalization, and J. Minnier performed statistical analysis. Q. Guo and P. Schedin wrote the manuscript with the help of J. Minnier and J. Burchard for the RNA-Seq methods section.
Supplementary Material
Acknowledgments
We acknowledge Ann Chambers (London Health Sciences Centre) and Jeffrey Green (National Cancer Institute) for providing the D2A1 tumor cell line, Peter Henson (National Jewish Health) for providing the L929 cell line, and Jun Xu, Tatiana Kisseleva, and David Brenner (University of California San Diego) for providing the pCol9GFP transgenic mouse mammary tissue. We thank Sushil Kumar (OHSU), Jaime Fornetti (University of Utah), and Traci Lyons (University of Colorado), and Schedin lab members Courtney Betts, Erica Goddard, Sonali Jindal, Nathan Pennock, and Elizabeth Mitchell for intellectual contributions. We are grateful to Jacob Fisher, Hadley Holden, Marcelia Brown, Jing Wang and Jayasri Narasimhan for technical assistance with animal husbandry and IHC. We also thank Lynn Sakai (OHSU) for critical review of the manuscript. In addition, we acknowledge the Flow Cytometry and Genomics and Microarray Cores at the University of Colorado Cancer Center (supported in part by Colorado’s NIH/NCI Cancer Center Support Grant P30CA046934) and the OHSU Flow Cytometry and Gene Profiling Shared Resources at Knight Cancer Institute Cancer Center (supported in part by OHSU’s NIH/NCI Cancer Center Support Grant P30CA069533). We thank Motifolio.com for providing predesigned diagrams for our figures. This work was supported by an OHSU Knight Cancer Institute sponsored graduate student stipend (to Q. Guo), the Department of Defense (W81XWH-11-1-0776 to P. Schedin), NIH/National Cancer Institute (1R01CA169175 to P. Schedin and V. Borges) and the Kay Yow Cancer Fund to P. Schedin.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(6):e89206. https://doi.org/10.1172/jci.insight.89206.
Contributor Information
Qiuchen Guo, Email: guoq@ohsu.edu.
Jessica Minnier, Email: minnier@ohsu.edu.
Julja Burchard, Email: burchard@ohsu.edu.
Kami Chiotti, Email: chiotti@ohsu.edu.
Paul Spellman, Email: spellmap@ohsu.edu.
Pepper Schedin, Email: schedin@ohsu.edu.
References
- 1.Stensheim H, Møller B, van Dijk T, Fosså SD. Cause-specific survival for women diagnosed with cancer during pregnancy or lactation: a registry-based cohort study. J Clin Oncol. 2009;27(1):45–51. doi: 10.1200/JCO.2008.17.4110. [DOI] [PubMed] [Google Scholar]
- 2.Johansson AL, Andersson TM, Hsieh CC, Cnattingius S, Lambe M. Increased mortality in women with breast cancer detected during pregnancy and different periods postpartum. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1865–1872. doi: 10.1158/1055-9965.EPI-11-0515. [DOI] [PubMed] [Google Scholar]
- 3.Callihan EB, et al. Postpartum diagnosis demonstrates a high risk for metastasis and merits an expanded definition of pregnancy-associated breast cancer. Breast Cancer Res Treat. 2013;138(2):549–559. doi: 10.1007/s10549-013-2437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amant F, et al. Prognosis of women with primary breast cancer diagnosed during pregnancy: results from an international collaborative study. J Clin Oncol. 2013;31(20):2532–2539. doi: 10.1200/JCO.2012.45.6335. [DOI] [PubMed] [Google Scholar]
- 5.Lyons TR, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat Med. 2011;17(9):1109–1115. doi: 10.1038/nm.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons TR, et al. Cyclooxygenase-2-dependent lymphangiogenesis promotes nodal metastasis of postpartum breast cancer. J Clin Invest. 2014;124(9):3901–3912. doi: 10.1172/JCI73777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinson HA, Jindal S, Durand-Rougely C, Borges VF, Schedin P. Wound healing-like immune program facilitates postpartum mammary gland involution and tumor progression. Int J Cancer. 2015;136(8):1803–1813. doi: 10.1002/ijc.29181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund LR, et al. Two distinct phases of apoptosis in mammary gland involution: proteinase-independent and -dependent pathways. Development. 1996;122(1):181–193. doi: 10.1242/dev.122.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schedin P, Mitrenga T, McDaniel S, Kaeck M. Mammary ECM composition and function are altered by reproductive state. Mol Carcinog. 2004;41(4):207–220. doi: 10.1002/mc.20058. [DOI] [PubMed] [Google Scholar]
- 10.Clarkson RW, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6(2):R92–109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein T, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6(2):R75–R91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien J, et al. Alternatively activated macrophages and collagen remodeling characterize the postpartum involuting mammary gland across species. Am J Pathol. 2010;176(3):1241–1255. doi: 10.2353/ajpath.2010.090735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jindal S, et al. Postpartum breast involution reveals regression of secretory lobules mediated by tissue-remodeling. Breast Cancer Res. 2014;16(2):R31. doi: 10.1186/bcr3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson CJ. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res. 2006;8(2):203. doi: 10.1186/bcr1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillitzer R, Goebeler M. Chemokines in cutaneous wound healing. J Leukoc Biol. 2001;69(4):513–521. [PubMed] [Google Scholar]
- 16.Midwood KS, Williams LV, Schwarzbauer JE. Tissue repair and the dynamics of the extracellular matrix. Int J Biochem Cell Biol. 2004;36(6):1031–1037. doi: 10.1016/j.biocel.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Wilgus TA. Immune cells in the healing skin wound: influential players at each stage of repair. Pharmacol Res. 2008;58(2):112–116. doi: 10.1016/j.phrs.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 19.Schäfer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9(8):628–638. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- 20.Montesano R, Orci L. Transforming growth factor beta stimulates collagen-matrix contraction by fibroblasts: implications for wound healing. Proc Natl Acad Sci USA. 1988;85(13):4894–4897. doi: 10.1073/pnas.85.13.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 22.Dumont N, et al. Breast fibroblasts modulate early dissemination, tumorigenesis, and metastasis through alteration of extracellular matrix characteristics. Neoplasia. 2013;15(3):249–262. doi: 10.1593/neo.121950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev. 2016;30(9):1002–1019. doi: 10.1101/gad.279737.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Threatt B, Norbeck JM, Ullman NS, Kummer R, Roselle P. Association between mammographic parenchymal pattern classification and incidence of breast cancer. Cancer. 1980;45(10):2550–2556. doi: 10.1002/1097-0142(19800515)45:10<2550::AID-CNCR2820451013>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Martin LJ, Boyd NF. Mammographic density. Potential mechanisms of breast cancer risk associated with mammographic density: hypotheses based on epidemiological evidence. Breast Cancer Res. 2008;10(1):201. doi: 10.1186/bcr1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conklin MW, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178(3):1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provenzano PP, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. Stiff collagen matrices increase tumorigenic prolactin signaling in breast cancer cells. J Biol Chem. 2013;288(18):12722–12732. doi: 10.1074/jbc.M112.447631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4(1):38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Brien J, et al. Non-steroidal anti-inflammatory drugs target the pro-tumorigenic extracellular matrix of the postpartum mammary gland. Int J Dev Biol. 2011;55(7–9):745–755. doi: 10.1387/ijdb.113379jo. [DOI] [PubMed] [Google Scholar]
- 33.Goswami S, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65(12):5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 34.Bates GJ, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24(34):5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- 35.Gao D, et al. Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res. 2012;72(6):1384–1394. doi: 10.1158/0008-5472.CAN-11-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiser J, Banerjee A. Effector, memory, and dysfunctional CD8(+) T cell fates in the antitumor immune response. J Immunol Res. 2016;2016:8941260. doi: 10.1155/2016/8941260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali HR, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14(2):R48. doi: 10.1186/bcr3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, et al. CD4+ and CD8+ T cells have opposing roles in breast cancer progression and outcome. Oncotarget. 2015;6(19):17462–17478. doi: 10.18632/oncotarget.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallina G, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116(10):2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer. 2016:2. doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward R, et al. Monocytes and macrophages, implications for breast cancer migration and stem cell-like activity and treatment. Oncotarget. 2015;6(16):14687–14699. doi: 10.18632/oncotarget.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silzle T, Kreutz M, Dobler MA, Brockhoff G, Knuechel R, Kunz-Schughart LA. Tumor-associated fibroblasts recruit blood monocytes into tumor tissue. Eur J Immunol. 2003;33(5):1311–1320. doi: 10.1002/eji.200323057. [DOI] [PubMed] [Google Scholar]
- 44.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17(2):135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 45.Yata Y, et al. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology. 2003;37(2):267–276. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 46.Vardell E. JoVE: the Journal of Visualized Experiments. Med Ref Serv Q. 2015;34(1):88–97. doi: 10.1080/02763869.2015.986795. [DOI] [PubMed] [Google Scholar]
- 47.Mosunjac MB, Lewis MM, Lawson D, Cohen C. Use of a novel marker, calponin, for myoepithelial cells in fine-needle aspirates of papillary breast lesions. Diagn Cytopathol. 2000;23(3):151–155. doi: 10.1002/1097-0339(200009)23:3<151::AID-DC2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 48.Stein T, Salomonis N, Nuyten DS, van de Vijver MJ, Gusterson BA. A mouse mammary gland involution mRNA signature identifies biological pathways potentially associated with breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2009;14(2):99–116. doi: 10.1007/s10911-009-9120-1. [DOI] [PubMed] [Google Scholar]
- 49.O’Brien JH, Vanderlinden LA, Schedin PJ, Hansen KC. Rat mammary extracellular matrix composition and response to ibuprofen treatment during postpartum involution by differential GeLC-MS/MS analysis. J Proteome Res. 2012;11(10):4894–4905. doi: 10.1021/pr3003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertolami C, Donoff RB. The effect of full-thickness skin grafts on the actomyosin content of contracting wounds. J Oral Surg. 1979;37(7):471–476. [PubMed] [Google Scholar]
- 51.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184(3):1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karin N. The multiple faces of CXCL12 (SDF-1alpha) in the regulation of immunity during health and disease. J Leukoc Biol. 2010;88(3):463–473. doi: 10.1189/jlb.0909602. [DOI] [PubMed] [Google Scholar]
- 53.Chatterjee M, et al. Platelet-derived CXCL12 regulates monocyte function, survival, differentiation into macrophages and foam cells through differential involvement of CXCR4-CXCR7. Cell Death Dis. 2015;6:e1989. doi: 10.1038/cddis.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su G, Sung KE, Beebe DJ, Friedl A. Functional screen of paracrine signals in breast carcinoma fibroblasts. PLoS ONE. 2012;7(10):e46685. doi: 10.1371/journal.pone.0046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esbona K, et al. COX-2 modulates mammary tumor progression in response to collagen density. Breast Cancer Res. 2016;18(1):35. doi: 10.1186/s13058-016-0695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. US Food and Drug Administration. Guidance for Industry. Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers 2005. http://www.fda.gov/ohrms/dockets/98fr/02d-0492-gdl0002.pdf Accessed Februrary 16, 2017.
- 57.Seluanov A, Vaidya A, Gorbunova V. Establishing primary adult fibroblast cultures from rodents. J Vis Exp. 2010;(44):2033. doi: 10.3791/2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakagawa S, Pawelek P, Grinnell F. Long-term culture of fibroblasts in contracted collagen gels: effects on cell growth and biosynthetic activity. J Invest Dermatol. 1989;93(6):792–798. doi: 10.1111/1523-1747.ep12284425. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen AV, Pollard JW. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Development. 2000;127(14):3107–3118. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 60.Gorska AE, Jensen RA, Shyr Y, Aakre ME, Bhowmick NA, Moses HL. Transgenic mice expressing a dominant-negative mutant type II transforming growth factor-beta receptor exhibit impaired mammary development and enhanced mammary tumor formation. Am J Pathol. 2003;163(4):1539–1549. doi: 10.1016/S0002-9440(10)63510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts AB, Sporn MB. Transforming growth factor-beta: potential common mechanisms mediating its effects on embryogenesis, inflammation-repair, and carcinogenesis. Int J Rad Appl Instrum B. 1987;14(4):435–439. doi: 10.1016/0883-2897(87)90020-1. [DOI] [PubMed] [Google Scholar]
- 62.Uría JA, Jiménez MG, Balbín M, Freije JM, López-Otín C. Differential effects of transforming growth factor-beta on the expression of collagenase-1 and collagenase-3 in human fibroblasts. J Biol Chem. 1998;273(16):9769–9777. doi: 10.1074/jbc.273.16.9769. [DOI] [PubMed] [Google Scholar]
- 63.Lijnen P, Petrov V, Rumilla K, Fagard R. Transforming growth factor-beta 1 promotes contraction of collagen gel by cardiac fibroblasts through their differentiation into myofibroblasts. Methods Find Exp Clin Pharmacol. 2003;25(2):79–86. doi: 10.1358/mf.2003.25.2.723680. [DOI] [PubMed] [Google Scholar]
- 64.Dewland PM, Reader S, Berry P. Bioavailability of ibuprofen following oral administration of standard ibuprofen, sodium ibuprofen or ibuprofen acid incorporating poloxamer in healthy volunteers. BMC Clin Pharmacol. 2009;9:19. doi: 10.1186/1472-6904-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyd NF, Martin LJ, Yaffe MJ, Minkin S. Mammographic density and breast cancer risk: current understanding and future prospects. Breast Cancer Res. 2011;13(6):223. doi: 10.1186/bcr2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fornetti J, Jindal S, Middleton KA, Borges VF, Schedin P. Physiological COX-2 expression in breast epithelium associates with COX-2 levels in ductal carcinoma in situ and invasive breast cancer in young women. Am J Pathol. 2014;184(4):1219–1229. doi: 10.1016/j.ajpath.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gillette M, Bray K, Blumenthaler A, Vargo-Gogola T. P190B RhoGAP overexpression in the developing mammary epithelium induces TGFβ-dependent fibroblast activation. PLoS ONE. 2013;8(5):e65105. doi: 10.1371/journal.pone.0065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blackstock CD, et al. Insulin-like growth factor-1 increases synthesis of collagen type I via induction of the mRNA-binding protein LARP6 expression and binding to the 5′ stem-loop of COL1a1 and COL1a2 mRNA. J Biol Chem. 2014;289(11):7264–7274. doi: 10.1074/jbc.M113.518951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takabatake Y, et al. Lactation opposes pappalysin-1-driven pregnancy-associated breast cancer. EMBO Mol Med. 2016;8(4):388–406. doi: 10.15252/emmm.201606273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDaniel SM, et al. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol. 2006;168(2):608–620. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huo CW, et al. High mammographic density is associated with an increase in stromal collagen and immune cells within the mammary epithelium. Breast Cancer Res. 2015;17:79. doi: 10.1186/s13058-015-0592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Acerbi I, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb) 2015;7(10):1120–1134. doi: 10.1039/C5IB00040H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes K, Wickenden JA, Allen JE, Watson CJ. Conditional deletion of Stat3 in mammary epithelium impairs the acute phase response and modulates immune cell numbers during post-lactational regression. J Pathol. 2012;227(1):106–117. doi: 10.1002/path.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cotechini T, Medler TR, Coussens LM. Myeloid cells as targets for therapy in solid tumors. Cancer J. 2015;21(4):343–350. doi: 10.1097/PPO.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, et al. Low/negative expression of PDGFR-α identifies the candidate primary mesenchymal stromal cells in adult human bone marrow. Stem Cell Reports. 2014;3(6):965–974. doi: 10.1016/j.stemcr.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bartol A, Laib A, Augustin H. Tumor vessel associated-pericytes. In: Mueller MM, NE Fusenig, eds. Tumor-Associated Fibroblasts and their Matrix. New York City, NY: Springer; 2011:91–97. [Google Scholar]
- 78.Viski C, König C, Kijewska M, Mogler C, Isacke CM, Augustin HG. Endosialin-expressing pericytes promote metastatic dissemination. Cancer Res. 2016;76(18):5313–5325. doi: 10.1158/0008-5472.CAN-16-0932. [DOI] [PubMed] [Google Scholar]
- 79.Palucka AK, Coussens LM. The basis of oncoimmunology. Cell. 2016;164(6):1233–1247. doi: 10.1016/j.cell.2016.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59(19):5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Connell JT, et al. VEGF-A and tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc Natl Acad Sci USA. 2011;108(38):16002–16007. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elkabets M, et al. Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J Clin Invest. 2011;121(2):784–799. doi: 10.1172/JCI43757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pazolli E, Alspach E, Milczªrek A, Prior J, Piwnica-Worms D, Stewart SA. Chromatin remodeling underlies the senescence-associated secretory phenotype of tumor stromal fibroblasts that supports cancer progression. Cancer Res. 2012;72(9):2251–2261. doi: 10.1158/0008-5472.CAN-11-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Özdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kwan ML, Habel LA, Slattery ML, Caan B. NSAIDs and breast cancer recurrence in a prospective cohort study. Cancer Causes Control. 2007;18(6):613–620. doi: 10.1007/s10552-007-9003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim S, et al. Lifetime use of nonsteroidal anti-inflammatory drugs and breast cancer risk: results from a prospective study of women with a sister with breast cancer. BMC Cancer. 2015;15:960. doi: 10.1186/s12885-015-1979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64(3):479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 88.Francke A, Herold J, Weinert S, Strasser RH, Braun-Dullaeus RC. Generation of mature murine monocytes from heterogeneous bone marrow and description of their properties. J Histochem Cytochem. 2011;59(9):813–825. doi: 10.1369/0022155411416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 90.Reddy SM, et al. Phagocytosis of apoptotic cells by macrophages induces novel signaling events leading to cytokine-independent survival and inhibition of proliferation: activation of Akt and inhibition of extracellular signal-regulated kinases 1 and 2. J Immunol. 2002;169(2):702–713. doi: 10.4049/jimmunol.169.2.702. [DOI] [PubMed] [Google Scholar]
- 91.Helft J, et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42(6):1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.