Conspectus
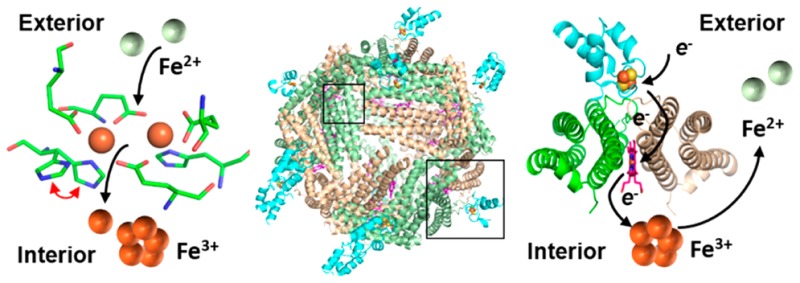
Despite its essentiality to life, iron presents significant challenges to cells: the exceedingly low solubility of Fe3+ limits its bioavailability, and the reactivity of Fe2+ toward H2O2 is a source of the toxic hydroxyl radical (HO•). Consequently, cellular levels of free iron are highly regulated to ensure sufficiency while preventing iron-induced toxicity. Relatively little is known about the fate of iron in the bacterial cytosol or how cells balance the need for relatively high cytosolic iron concentrations with the potential toxicity of the nutrient. Iron storage proteins are integral to iron metabolism, and bacteria utilize two types of ferritin-like molecules to store iron, bacterial ferritin (Ftn) and bacterioferritin (Bfr). Ftn and Bfr compartmentalize iron at concentrations far above the solubility of Fe3+ and protect the reducing cell environment from unwanted Fe3+/Fe2+ redox cycling.
This Account focuses on our laboratory’s efforts to study iron storage proteins in the model bacterium Pseudomonas aeruginosa, an opportunistic pathogen. Prior to our studies, it was thought that P. aeruginosa cells relied on a single Bfr assembled from two distinct subunits coded by the bfrA and bfrB genes. It is now known that, like in most bacteria, two iron storage proteins coexist in P. aeruginosa cells, a bacterial Ftn (FtnA), coded by the ftnA (formerly bfrA) gene and a bacterioferritin (BfrB), coded by the bfrB gene. Studies with BfrB showed that Fe2+ oxidation occurs at ferroxidase centers (FCs), followed by gated translocation of Fe3+ to the interior cavity, a process that is, surprisingly, distinct from that observed with the extensively studied Bfr from Escherichia coli, where the FCs are stable and function only as a catalytic site for O2 reduction. Investigations with BfrB showed that the oxidation of Fe2+ at FCs and the internalization of Fe3+ depend on long-range cooperative motions, extending from 4-fold pores, via B-pores, into FCs. It remains to be seen whether similar studies with E. coli Bfr will reveal distinct cooperative motions contributing to the stability of its FCs. Mobilization of Fe3+ stored in BfrB requires interaction with a ferredoxin (Bfd), which transfers electrons to reduce Fe3+ in the internal cavity of BfrB for subsequent release of Fe2+. The structure of the BfrB/Bfd complex furnished the only known structure of a ferritin molecule in complex with a physiological protein partner. The BfrB/Bfd complex is stabilized by hot-spot residues in both proteins, which interweave into a highly complementary hot region. The hot-spot residues are conserved in the sequences of Bfr and Bfd proteins from a number of bacteria, indicating that the BfrB/Bfd interaction is of widespread significance in bacterial iron metabolism. The BfrB/Bfd structure also furnished the only known structure of a Bfd, which revealed a novel helix-turn-helix fold different from the β-strand and α-helix fold of plant and vertebrate [2Fe–2S]-ferredoxins. Bfds seem to be unique to bacteria; consequently, although mobilization of iron from eukaryotic ferritins may also be facilitated by protein–protein interactions, the nature of the protein that delivers electrons to the ferric core of eukaryotic ferritins remains unknown.
Introduction
Iron in biological systems occurs predominately in cofactors such as heme, iron centers, and iron–sulfur clusters, which are integral to enzymes that function in critical processes, such as O2 transport and activation, respiration, DNA synthesis, and gene regulation. The predominance of iron in biological systems presumably arose because of its large abundance on the Earth’s crust when an O2-depleted atmosphere facilitated the aqueous solubility of Fe2+. The rise of atmospheric O2 caused oxidation to the insoluble Fe3+, drastically decreasing bioavailability and also creating the potential for iron-induced toxicity when intracellular iron and O2 react to produce reactive oxygen species (O2•–, H2O2, and HO•).1,2 To ensure iron sufficiency while preventing iron-induced toxicity, organisms maintain iron homeostasis by balancing iron scavenging with iron storage and utilization. Particular to iron homeostasis are the ferritins,3,4 which function by utilizing O2 or H2O2 to oxidize Fe2+ and by compartmentalizing the resultant Fe3+.
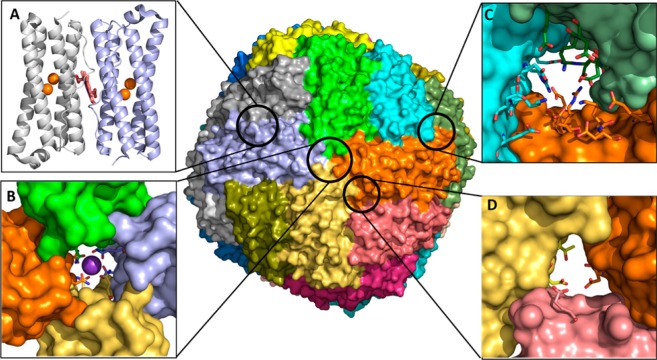
Bacteria have two types of ferritin-like molecules, the bacterial ferritins (Ftn) and the bacterioferritins (Bfr).3,4 Ftns and Bfrs assemble from 24 subunits into spherical hollow structures (∼120 Å outer diameter, ∼80 Å inner diameter) (Figure 1). Each subunit consists of a four-helix bundle with a loop connecting helices B and C and a short C-terminal α-helix perpendicular to the bundle (Figure 1A). Despite similar fold and quaternary structure, bacterial Ftns, Bfrs, and eukaryotic Ftns differ significantly:
Low sequence similarity (<20%) causes distinct subunit packing, charge distribution, and possibly function.5−10
The 24-mer eukaryotic Ftns assemble from two distinct subunits (H and L); only the H subunits harbor catalytic ferroxidase centers (FCs) for the oxidation of Fe2+ to Fe3+,8−10 whereas 24-mer bacterial Ftns and Bfrs assemble from identical subunits, each harboring a FC.3,4
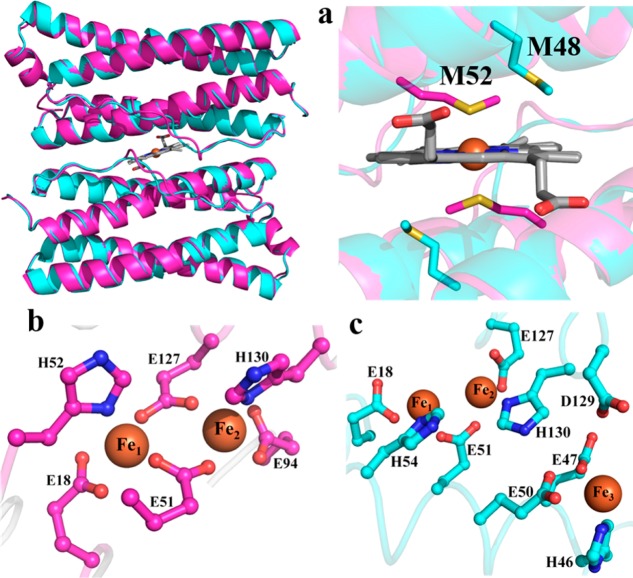
Only Bfrs bind heme at 2-fold intersubunit sites where each heme-iron is coordinated by a conserved M52 from each subunit (Figure 1A).3,4,11
All 24-mer Ftns have eight 3-fold and six 4-fold pores, but the composition and electrostatic properties of the pores vary significantly. The 3-fold pores of Ftns and Bfrs are lined by alternating layers of positively and negatively charged residues, E109, R117, K121, and D122 in BfrB (Figures 1C and 8). The 4-fold pores are lined with hydrophilic residues N148 and Q151 (Figure 1B); in most crystal structures these residues bind a monovalent or divalent cation other than iron (Figure 1B).
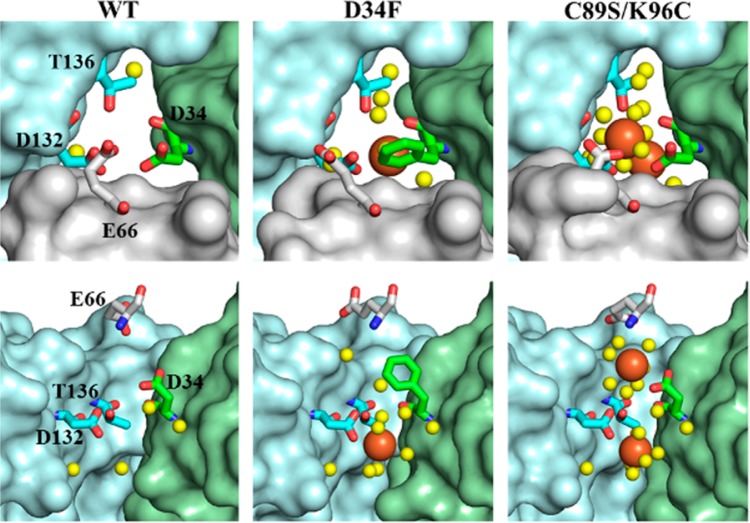
Only bacterial Ftns and Bfrs have B-pores, which are formed at an asymmetric site between three subunits and are lined with hydrophilic and negatively charged residues, D34, E66, D132, and T136 in BfrB (Figures 1D and 7).
Figure 1.
BfrB is a nearly spherical molecule assembled from 24 identical subunits and 12 hemes. (A) Each subunit harbors a FC, and each heme is at the interface of 2 subunits; iron in the FCs is shown as orange spheres. The interior cavity is in contact with the exterior via 4-fold pores (B) (K+ present in each of the 4-fold pores is shown as a purple sphere), 3-fold pores (C), and B-pores (D).
Figure 8.
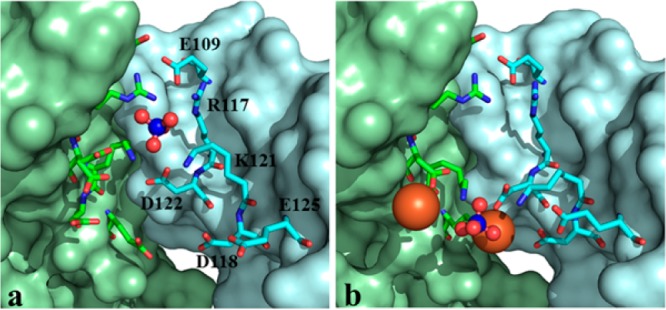
Comparing the 3-fold pores in as isolated (a) and Fe-soaked (b) C89S/K96C BfrB suggest a possible path for the access of phosphate to the interior cavity where they encounter Fe3+. Fe ions are shown in orange and sulfate in red and blue spheres.
Figure 7.
B-pores in Fe-soaked structures viewed from the protein exterior (top) and from the side with the gray subunit removed (bottom). Iron ions are in orange and water in yellow spheres.
The growing realization of the differences between Ftns, Bfrs, and eukatyotic ferritins, which stems from the characterization of an increasing number of molecules from distinct organisms, is fueling a mounting awareness that despite very similar structural architectures, the differences probably originate from evolutionary adaptations that enable organisms to succeed in their environmental niche.7 These adaptations may occur via mutations that maintain overall structure but impart mechanistic changes, which in turn endow 24-mer Ftns with primary functions that are not necessarily iron storage, for example, antioxidant or stress sensing.7,8 In this context, the aerobic oxidative accumulation of Fe2+ by Bfr from Pseudomonas aeruginosa (BfrB),12 which was found to occur differently than in the widely studied Escherichia coli Bfr,7 despite very similar structures, constitutes the first clear example that subtle structural differences indeed affect mechanism. Efforts aimed at understanding the mechanism of oxidative accumulation of Fe2+ by BfrB in the context of structure and long-range dynamic communication within the 24-mer are described here. This Account also summarizes the molecular-level description of the BfrB/Bfd complex, which functions in the mobilization of iron from BfrB and represents the first and so far unique structure of a protein–protein complex involving a Ftn or Ftn-like molecule.
Two Types of Ferritin-Like Molecule Coexist in P. aeruginosa Cells, FtnA and BfrB
Pioneering studies conducted with Bfr isolated from P. aeruginosa suggested that the protein is assembled from two distinct subunits, akin to vertebrate Ftns.13 Subsequent investigations suggested two distinct genes coding for bacterioferritin in P. aeruginosa, bfrA and bfrB. Interrogation of the global genetic response of P. aeruginosa to high and low iron conditions showed that the bfrB gene is upregulated in response to high iron conditions, whereas expression of bfrA does not respond to external iron concentrations.14,15 Our sequence alignments showed that M52, the conserved heme-ligand in Bfrs, is absent in the sequence encoded by the bfrA gene, leading us to hypothesize that bfrA encodes a bacterial Ftn (not a Bfr) and to suggest that the products of the bfrA and bfrB genes assemble into distinct molecules.12 Recombinant expression of bfrA and bfrB allowed us to show that the corresponding products indeed assemble into independent 24-mer proteins, and our structural work demonstrated that although the subunits in each protein adopt an identical fold (Figure 2a) and assemble into 24-mer proteins with similar quaternary structure, an obvious distinction is that the protein coded by bfrB binds heme, whereas the protein coded by bfrA does not.12,16 In a BfrB subunit dimer, M52 is located at the center of the B-helix and is ideally positioned to coordinate the heme-iron. In contrast, M48 in BfrA is too far away to bind the heme-iron, demonstrating that BfrA did not evolve to bind heme. BfrA and BfrB also differ in the structure of their FCs. The FCs in BfrB are typical of Bfrs,3 with Fe1 and Fe2 coordinated by two bridging glutamates and capped by Glu and His ligands (Figure 2b). In contrast, the FCs in BfrA are similar to the FCs of bacterial Ftns, where only one glutamate bridges Fe1 and Fe2, and a third iron site (Fe3) is also present. Given these structural attributes, it is clear that the protein coded by the bfrB gene (BfrB) is a genuine bacterioferritin,12 whereas the protein coded by the bfrA gene is a bacterial ferritin, which we proposed should be termed FtnA.16 The Pseudomonas Genome Database (www.pseudomonas.com) now lists gene PA4235 (formerly bfrA) as ftnA and gene PA3531 as bfrB.
Figure 2.
(a) Superposition of two subunits in BfrB (magenta) with two subunits in FtnA (cyan); the zoomed-in view of the heme binding site in BfrB shows that M48 in FtnA cannot bind heme. The FCs in BfrB (b) are different from those in FtnA (c), which are structurally related to FCs in bacterial Ftns.
In P. aeruginosa, the ftnA gene is expressed constitutively and is adjacent to katA, which codes for a heme catalase (KatA).17 Interestingly, a ftnA-null mutant of P. aeruginosa showed 50% of the catalase activity of wild type cells, which led to the speculation that iron stored in FtnA (formerly BfrA) is incorporated in protoporphyrin IX to make the heme cofactor of KatA.17 In comparison, transcription of the bfrB gene is stimulated by iron, and bfrB is located adjacent to bfd, which codes for Bfd, a bacterioferritin-associated ferredoxin. Early studies suggested that E. coli Bfr and Bfd form an electron transfer complex, which was postulated to participate in the iron storage or in the iron mobilization functions of Bfr.18,19 These observations prompted us to carry out detailed studies of P. aeruginosa BfrB, Bfd, and the BfrB/Bfd complex, which demonstrated that the electron transfer complex functions in the mobilization of iron stored in BfrB (see below).20−22
Iron Oxidation and Uptake by BfrB
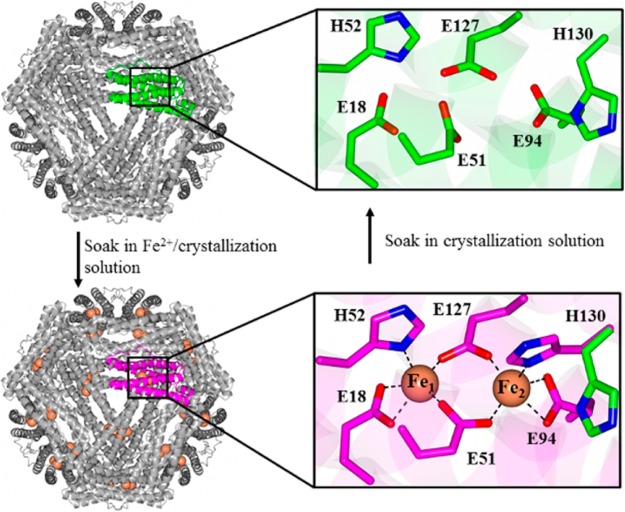
The BfrB structure was solved using crystals grown from (i) BfrB devoid of iron (as-isolated), (ii) BfrB reconstituted with 600 iron atoms (mineralized), (iii) crystals of mineralized BfrB after soaking in crystallization solution containing FeSO4 (Fe-soaked), and (iv) crystals of Fe-soaked BfrB after soaking in crystallization solution (double-soaked). The BfrB structures suggested possible paths for admission of Fe2+ into the FCs and, after oxidation, for translocation of Fe3+ from the FC to the interior cavity:12 The FCs in as-isolated BfrB are devoid of iron, and although most of the FC ligands are poised to bind iron, the side chains of H130 are rotated away (Figure 3 top). To promote iron binding at the FCs, BfrB was reacted with Fe2+ and purified with an iron core of 600 Fe3+ ions (mineralized) prior to crystal growth. Despite having processed 600 iron ions, the FCs of mineralized BfrB remained empty, with ferroxidase ligands adopting conformations identical to those seen in the as-isolated protein (Figure 3 top). In contrast, the Fe-soaked structure showed iron in the FCs (Figure 3 bottom) and revealed that the side chains of FC ligands undergo minimal reorganization upon iron binding, except H130, which was observed in two conformations, one poised to bind Fe2 (∼70% occupancy), termed “gate closed” and the second similar to that in empty FCs, termed “gate open”. When Fe-soaked crystals were soaked in crystallization solution, the FCs were empty, and the FC ligands adopted conformations identical to those in the as-isolated structure. We interpreted these observations to indicate that the FCs in BfrB function in a dual role of Fe2+ oxidizing center and pore for internalization of Fe3+.12 Key to this dual function is the flexibility of the H130 side chains, which rotate from a “gate open” conformation in empty FCs to a “gate closed” conformation to bind iron at di-Fe2+ centers. Subsequent oxidation to di-Fe3+ centers probably triggers a rearrangement to the “gate open” conformation, which allows translocation of Fe3+ from the FCs to the interior cavity.
Figure 3.
FCs in as isolated BfrB are empty (top). Soaking crystals of as isolated BfrB in Fe2+ solution allows observation of iron loaded FCs and of a conformational rearrangement of the H130 side chains from “gate open” (top) to “gate closed” (bottom). Soaking Fe-soaked crystals in crystallization solution causes the FCs to empty, with concomitant rearrangement of H130 from the “gate closed” to the “gate open” conformation.
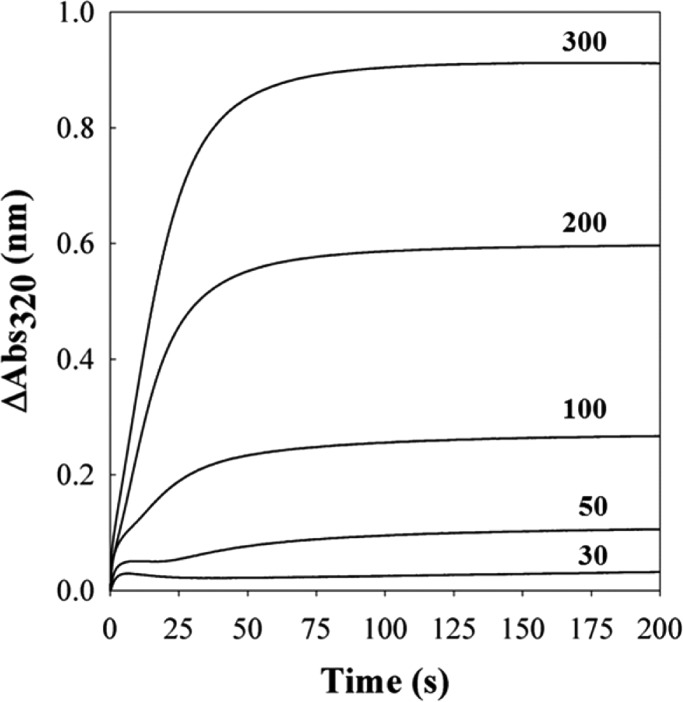
Additional evidence indicating that the FCs in BfrB function as catalytic sites for Fe2+ oxidation and pores for Fe3+ internalization was obtained using stopped-flow methods.12 When BfrB is mixed with solutions delivering <100 Fe2+/BfrB, the progress curves allow distinction of a fast phase lasting ∼10 s and a slower phase that becomes progressively faster as the iron load increases, such that when the load is >200 Fe2+/BfrB the two phases become indistinguishable (Figure 4). At iron loads of 30 and 50, the initial burst is followed by a decrease before the onset of the slower phase. The fast burst was interpreted to indicate oxidation of di-Fe2+ to di-Fe3+ centers and the subsequent decrease to indicate migration of Fe3+ from the FCs to the BfrB interior cavity, where mineralization (slow phase) takes place. This model is in agreement with that proposed for Fe2+ oxidation and uptake by vertebrate Ftns.8,10 It is interesting to note, however, that in E. coli Bfr iron forms a stable complex at the FCs, which continuously cycle between di-Fe2+ and di-Fe3+, driven by the oxidation of Fe2+ that gain entry to the interior cavity via pores in the Bfr structure.7 Hence, whereas the iron complexes in the FCs of BfrB are unstable and function as Fe2+ oxidation centers and pores for iron access, the iron complexes in FCs of E. coli Bfr are stable and function only as oxidation centers. These intriguing differences underscore the fact that in the complex architecture of 24-mer Bfrs, subtle structural differences exert profound influence on function, which may have been evolutionarily tailored to support adaptations of bacteria to their environmental niche. The influence exerted by subtle structural changes in the Bfr 24-mer assemblies is further illustrated below.
Figure 4.
Progress curves obtained upon mixing BfrB and iron solutions delivering 30, 50, 100, 200, and 300 Fe2+/BfrB.
BfrB Dynamics and Iron Uptake Function
Our normal-mode analysis of Ftn and Ftn-like molecules showed extensive long-range communication between ferroxidase centers and specific pores in the corresponding structures.6 In bullfrog Ftn M, a network of correlated residues connects FCs with 3-fold pores, whereas in bacterial Ftn and Bfr, networks of correlated residues connect the FCs with 4-fold and B-pores.6 The differences in long-range cooperativity between eukaryotic Ftn and bacterial Ftn and Bfr are probably a consequence of differences in quaternary packing, underscoring once again that subtle differences in the 24-mer assemblies profoundly affect function.
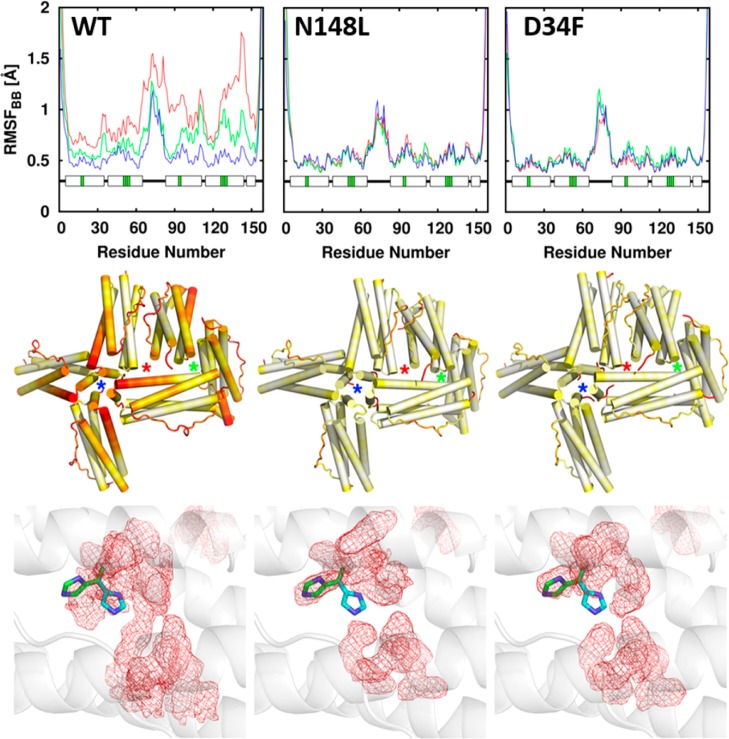
Atomistic molecular dynamics (MD) simulations conducted with wt BfrB revealed fluctuations coupling FCs with 4-fold- and B-pores, which appear to be important for ion traffic across the BfrB shell through the B-pores.23 Plots of per-residue root-mean square fluctuations (RMSFs) (Figure 5 top) showed that the C-terminal half of helix D exhibits the highest flexibility and that the flexibility of wt BfrB increases with ionic strength. Visualizing this pattern in the context of a 24-mer assembly (Figure 5 middle) reveals that in wt BfrB regions of high flexibility cluster around the 4-fold and B-pores, while regions comprising the 3-fold pores exhibit much less dynamical activity. To probe the significance of the networked motions linking FCs and pores in BfrB, we prepared mutants designed to lower the flexibility of the pores. N148L was prepared to replace the side chains that coordinate K+ in the 4-fold pores (see Figure 1) with a hydrophobic residue and D34F to replace a ligand known to coordinate K+ in the B-pores.24 X-ray diffraction showed only minor structural perturbations at the mutation sites, but MD simulations conducted with the corresponding coordinates showed significantly attenuated flexibility surrounding the 4-fold and B-pores in the mutants relative to wt BfrB (Figure 5 middle). Interestingly, the dampened folding/unfolding transitions in the C-terminal half of helices D also restrict the conformational flexibility of the H130 side chains. This is illustrated in the bottom panels of Figure 5, where it can be seen that in the wt MD simulations the average conformation of the H130 side chains (red mesh) is midway between the “gate open” (green sticks) and “gate closed” (cyan sticks) states. In contrast, the average conformation of H130 side chains in the N148 and D34F mutants aligns exclusively with the “gate open” conformation.24
Figure 5.
(top) Per-residue backbone RMSF; blue, green, and red plots correspond to increasing ionic strength, respectively. (middle) RMSF mapped onto six subunits of a 24-mer assembly encompassing 4-fold (blue stars), 3-fold (green stars), and B-pores (red stars) shows that the flexibility near 4-fold and B-pores in wt BfrB is dampened in the mutants. In the color scale, flexibility increases from white to red. (bottom) The average conformation explored by FC residues during MD simulations is shown as red mesh; the gate open (green) and gate closed (cyan) conformations of H130 are shown in sticks.
The FCs in as isolated WT, N148, and D34F BfrB are devoid of iron (Figure 6, left).24 The Fe-soaked structures, however, revealed striking differences: While iron loading onto the FCs of wt BfrB is accompanied by a rearrangement of H130 from “gate open” to “gate closed”, H130 in the FCs of D34F and N148L BfrB remains in the “gate open” conformation and E51 does not rotate to bridge Fe1 and Fe2 (Figure 6, right). These observations underscore the low flexibility of the FC ligands in the mutants and lend strong support to the idea that decreased flexibility in the pores also lowers the conformational agility of FC ligands. Importantly, the decreased flexibility of FC ligands in the mutant proteins, predicted by MD simulations and X-ray diffraction, also depresses the Fe2+ oxidation activity in solution.24 Taken together, the observations indicate that concerted motions connecting 4-fold and B-pores with FCs in BfrB are crucial to endow the FC ligands with the conformational freedom required to bind and oxidize di-Fe2+ centers and to gate the entry of Fe3+ to the interior cavity.6,23,24
Figure 6.
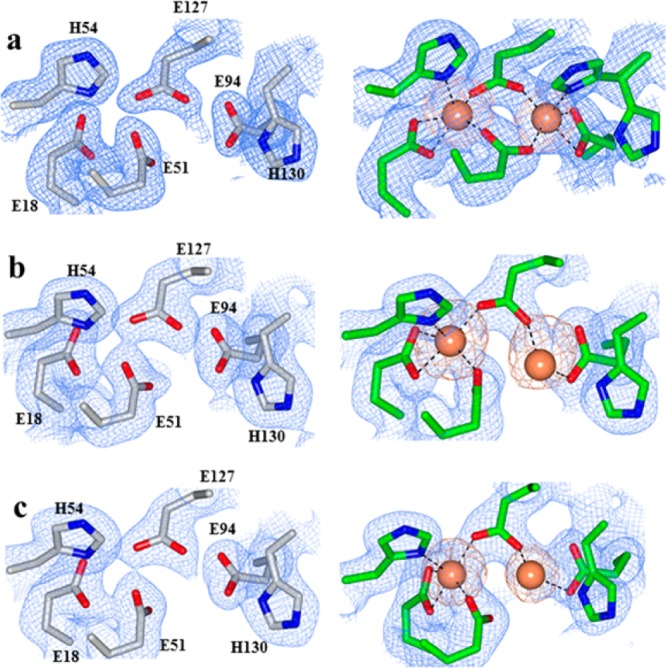
Electron density maps of FC ligands (blue mesh) and iron atoms (orange mesh) in (a) WT, (b) N148L, and (c) D34F BfrB. FCs from as isolated structures are on the left column and FCs from Fe-soaked structures on the right.
How Does Iron Traffic in and out of Bfr?
The interior of the 4-fold pores in Bfrs is lined with hydrophilic residues.3 In BfrB N148 and Q151 coordinate a K+ ion present in the 4-fold pores (Figure 1) in both as-isolated and Fe-soaked structures. Interestingly, although the 4-fold pores of Pa, Azotobacter vinelandii (Av), and Ec Bfr are structurally identical, metal ions other than Fe have been observed only in the 4-fold pores of Av and Pa BfrB.25,26 The B-pores in wt BfrB are demarcated by E66, D34, T136, and D132, arranged in a corkscrew (Figure 7). MD simulations conducted with wt BfrB showed that folding/unfolding transitions in the C-terminus of helix D enable K+ to traverse B-pores, suggesting that these channels may function as conduits for Fe2+ traffic.23 Experimental evidence for iron trafficking across B-pores was obtained in the crystal structure of D34F BfrB, a mutant prepared to restrict the dynamic motions of B-pores (see above).24 The as-isolated structure showed that F34 partially obstructs the B-pores and disrupts the hydrophilicity of the corkscrew, and the Fe-soaked structure revealed iron in the B-pores, coordinated by D132 (Figure 7). Additional evidence was obtained in the crystal structure of C89S/K96C BfrB, prepared to relocate the only surface cysteine (C89) for subsequent attachment of paramagnetic tags to the BfrB surface. The B-pores in as isolated C89S/K96C BfrB are structurally indistinguishable from those in wt BfrB, but the Fe-soaked structure shows two Fe ions aligned along the length of B-pores; the innermost coordinated by D132 and the outermost by D34 and E66 (Figure 7).24 It is interesting to note that studies with a D132F mutant of Ec Bfr, which is similar to D34F BfrB in that a hydrophobic residue substitutes a B-pore aspartate, showed decreased iron oxidation activity. These observations were interpreted to indicate that obstructing B-pores slows Fe2+ access to the interior cavity.27 It is also possible, however, that the decreased iron oxidation activity of the Ec D132F Bfr mutant is related to reduced conformational flexibility affecting the reactivity of the FCs, as has been seen with the D34F mutant in BfrB (see above).
Phosphate Traverses BfrB across 3-Fold Pores?
Unlike the core iron minerals of native eukaryotic Ftns, the core mineral of bacterial Ftns and Bfrs isolated from native sources contain high levels of phosphate (Fe:P ≈ 1:1).7,9 Consequently, iron uptake or mobilization from the Bfr cavity is probably accompanied by a corresponding flux of phosphate ions across the BfrB shell.3,12,24 In this context, it is noteworthy that although the 3-fold pores of as-isolated BfrB (wt and mutants) are empty, the Fe-soaked structures invariably reveal sulfate ions (from the crystallization solution) near the middle of the 3-fold pores, coordinated by R117 and K121. We have also been able to observe sulfate ion in the 3-fold pores of BfrB by prolonged soak of crystals of as isolated C89S/K96C BfrB in crystallization solution. The sulfate ions in this structure are also in the middle of the pore, coordinated by R117 and K121 (Figure 8a). When these crystals were soaked in Fe2+ solution, however, sulfate was observed at the bottom of the 3-fold pores, at the exit into the interior cavity, coordinated by the side chains of K121 and interacting with three iron ions (Figure 8b). We have observed a similar arrangement of Fe and SO42– in the Fe-soaked structures of N148L and D34F BfrB, and interpreted the observations to suggest that phosphate ions utilize the 3-fold pores to access the BfrB interior cavity where they encounter Fe3+ prior to being incorporated in the growing mineral.24
Global View of Iron in the BfrB Protein
Although the Fe-soaked structures of wt, N148L, and D34F BfrB revealed distinct iron binding sites in BfrB, the Fe-soaked structure of C89S/K96C BfrB allowed simultaneous observation of all the iron binding sites observed in the other structures, and a few previously unobserved.24Figure 9 shows a transverse view of the interior cavity in Fe-soaked C89S/K96C BfrB, with the 4-fold pores highlighted in purple, the 3-fold pores in blue, the B-pores in green, the ferroxidase pores in yellow, the iron ions in orange, and the phosphate ions in yellow and red. The view illustrates how iron can access the interior cavity via FCs and B-pores. Iron entering via FCs (probably Fe3+) can be transported to nearby 3-fold pores, where it may interact with PO43– accessing the cavity via these conduits. Iron entering via B-pores (probably Fe2+) can readily access nearby FCs for subsequent oxidation, as suggested by the MD simulations,23 or it can be oxidized by electron exchange with iron bound at FC sites, as proposed for Ec Bfr.7,28 It is noteworthy that iron has not yet been observed inside 4-fold pores, but several BfrB structures have consistently shown iron ions in the external perimeter of the 4-fold pores, so it is tempting to speculate that these sites function as binding sites prior to oxidation at FCs or incorporation into the iron mineral.24
Figure 9.

Transverse view of the interior cavity in Fe-soaked C89S/K96C BfrB, depicting iron ions in orange and sulfate ions in yellow and red spheres.
Protein–Protein Interactions Are Required To Mobilize Iron Stored in BfrB
Approximately 118 genes of the P. aeruginosa chromosome are upregulated in response to low iron conditions.14 We recognized that among these genes only bfd and fpr (ferredoxin NADP reductase) code for proteins with obvious electron transfer function and interpreted the observations to suggest a model for mobilizing Fe2+ from BfrB (Figure 10A), where Bfd mediates electrons from Fpr to BfrB.20,21 To challenge the model, we determined that the products of the bfd and fpr genes code for a [2Fe–2S]-ferredoxin (Bfd) and for an NADPH-dependent flavoenzyme (Fpr), respectively,29,30 and reconstituted the proteins with BfrB in vitro. Addition of NADPH to a solution containing mineralized BfrB, Bfd, and Fpr initiates the reactions shown in Figure 10A and causes the rapid and quantitative mobilization of iron from BfrB. If Bfd is omitted, however, NADPH does not promote iron mobilization from BfrB. Similar experiments conducted with FtnA instead of BfrB showed that Bfd is not necessary for iron mobilization from FtnA (Figure 10B),16 underscoring the idea that FtnA and BfrB most likely have distinct functions in P. aeruginosa.
Figure 10.
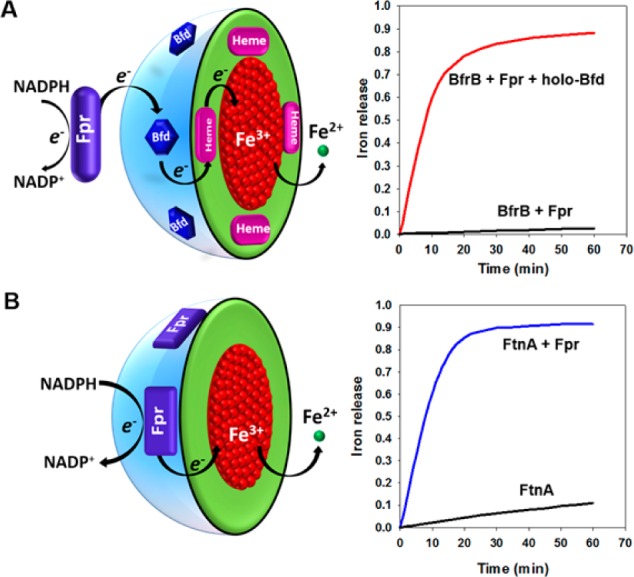
Mobilization of iron stored in BfrB requires Fpr and Bfd, whereas mobilization of iron stored in FtnA requires only Fpr.
The presence of heme in Bfr suggests that the cofactor transfers electrons across BfrB, either from the external surface to the ferric mineral for Fe2+ mobilization or from the core to the external surface for Fe2+ oxidation and Fe3+ uptake. Although evidence for the latter is thus far absent, it is now clear that heme mediates electrons from the surface to the mineral. The first indications came from observations of a biphasic heme reduction on addition of dithionite to iron-loaded Av-Bfr,31 and subsequently when experiments with Ec-BfrB showed that the rate of iron mobilization is faster when Ec-Bfr has five hemes/24-mer than when it contains only one heme/24-mer.32 Finally, UV–vis spectroscopy was used to demonstrate that the [2Fe–2S] cluster in Bfd transfers electrons to the heme in BfrB, which in turn transfers electrons to the Fe3+ core in BfrB.20,21
Structure of the BfrB/Bfd Complex
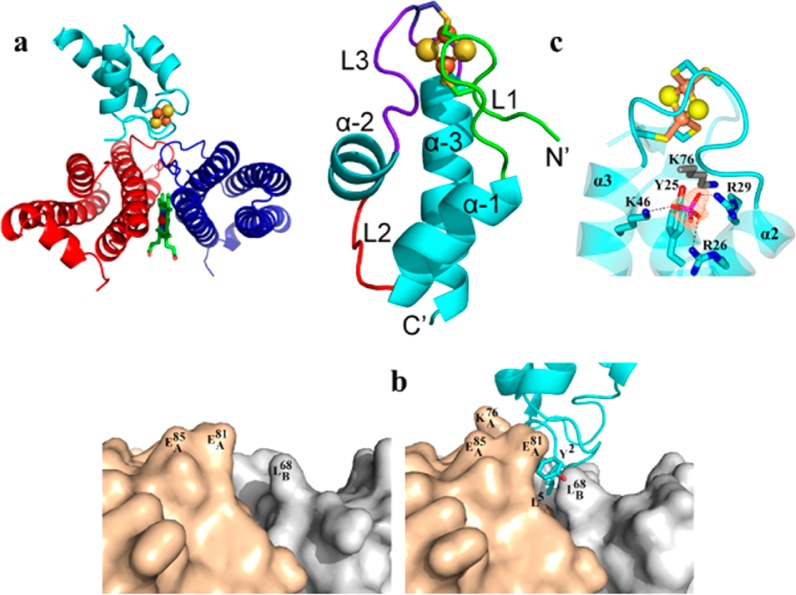
We solved the structure of the BfrB/Bfd complex at 2.0 Å resolution.21 It revealed a biological assembly consisting of 12 Bfd molecules bound to a 24-mer BfrB. Each Bfd binds at a structurally identical site on BfrB, at the interface of a subunit dimer, above a heme molecule (Figure 11a). Although Bfd binding triggers only small changes on the BfrB surface (reorientation of the L68, E81, and E85 side chains), these seemingly minor rearrangements are crucial to the stability of the BfrB/Bfd complex (see below): Reorientation of the L68 and E81 side chains on the BfrB surface narrows a cleft where the Y2 and L5 side chains of Bfd anchor (Figure 11b) and enables hydrogen bonding interactions between the carboxylate groups of E81 and E85 in BfrB and backbone groups of M1 and Y2 in Bfd (Figure 12a). The BfrB/Bfd structure also revealed the first structure of a Bfd molecule (Figure 11c). The Bfd fold consists of a helix-turn-helix fold, which is different from the β-strand covered by an α-helix fold typical of plant- and vertebrate-type [2Fe–2S]-ferredoxins. The [2Fe–2S] cluster is located in two nearly parallel hairpin loops, L1 and L3, which harbor ligands C4 and C6 and C38 and C41, respectively. A phosphate ion coordinated by R26, R29, and K46 appears to play an important role in the stability of the Bfd fold and the integrity of [2Fe–2S] cluster.21 Residues M1, Y2, and L5, which are located in L1, anchor Bfd at the BfrB surface. Consequently, in addition to electron transfer, the [2Fe–2S] cluster also functions to structure L1 and enable the side chains of M1, Y2, and L5 in Bfd to adopt the required conformations to effectively interact with the BfrB surface.
Figure 11.
Structure of the BfrB/Bfd complex showed that Bfd binds at the interface of two BfrB subunits (a) with minimal rearrangement of the BfrB surface (b) and also revealed a unique fold for the [2Fe–2S]-Bfd (c).
Figure 12.
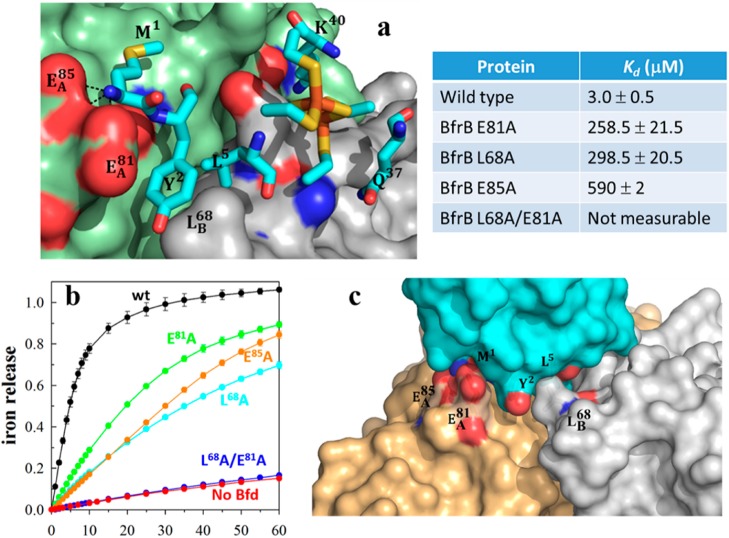
(a) Zoomed-in view of the BfrB/Bfd interface depicting the BfrB surface in green (subunit A) and gray (subunit B), residues anchoring Bfd as cyan sticks, and the [2Fe–2S] cluster in orange and yellow. (b) Perturbation of the cleft defined by L68 and E81 in BfrB, which abolishes binding to Bfd, also inhibits iron mobilization from BfrB. (c) Hot region of the BfrB/Bfd complex.
The BfrB/Bfd Complex Interface Is Conserved in Bacteria
With the aid of surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC), we showed that the 12 Bfd-binding sites on BfrB are equivalent and independent. Bfd binds to each of these sites with Kd = 3 μM, in an entropically driven process.22 Residues L68 and E81 on BfrB form a continuous set of interactions with M1, Y2, and L5 in Bfd (Figure 12a). Accordingly, the stability of the complex is highly sensitive to perturbations affecting L68 or E81. For instance, Kd values for the association between Bfd and E81A or L68A BfrB are 80- and 100-fold larger, respectively, than the Kd for the association with wt BfrB, and the Kd for the association between Bfd and L68A/E81A BfrB is too large to measure. In agreement with the lower binding affinities between Bfd and BfrB mutants, the rates of iron mobilization from L68A and E81A BfrB are slower than from wt BfrB, and iron mobilization from L68A/E81A BfrB is completely inhibited (Figure 12b).
Hot spot residues at protein–protein interfaces, which contribute to the majority of the binding energy, tend to occur in clusters, which often form an extensive set of interactions (hot region) at protein–protein interfaces.33 Our mutational analysis of the BfrB/Bfd interface identified L68, E81, and E85 in BfrB and Y2 and L5 in Bfd as hot spot residues. The view in Figure 12c illustrates how these residues interact to define a hot region. An important outcome of having identified the hot region stabilizing the BfrB/Bfd interaction is the realization that the hot spot residues at the interface are conserved in the sequences of Bfd and Bfr from a large number of Gram-negative bacteria, where the bfd and bfr genes are also adjacent in the corresponding chromosomes (see Table 3 in ref (22)). These observations strongly suggest that the BfrB/Bfd interaction is conserved in many bacteria and that the insights gained from studying BfrB and the BfrB/Bfd interaction in P. aeruginosa are likely of widespread significance in bacterial iron metabolism.21,22
Concluding Remarks
Recent discoveries are changing the long-held perception of Bfrs as “rigid cages” into highly tuned assemblies whose iron uptake/mobilization function depends strongly on long-range dynamic allosteric communication and protein–protein interactions. The now evident requirement of a ferredoxin (Bfd) to mobilize iron deposited in BfrB is the first demonstration of the participation of protein–protein interactions in the mobilization of iron stored in a ferritin or ferritin-like molecule. This discovery is expected to enable new interrogation of the role played by bacterioferritin as dynamic regulator of intracellular iron levels in bacteria.
Acknowledgments
The author thanks Drs. H. Yao, R. Kumar, and S. Lovell for helpful conversations. The author thanks NSF (MCB1615767) and NIH (AI125529) for support.
Biography
Mario Rivera received his Ph.D. in Analytical Chemistry from the University of Arizona, under the Direction of the late Prof. Quintus Fernando. He completed his postoctoral studies in Bioinorganic Chemistry at the University of Arizona, under the Direction of Prof. F. Ann Walker. He is currently a Professor of Chemistry at the University of Kansas.
The author declares no competing financial interest.
References
- Nielsen F. H. Evolutionary events culminating in specific minerals becoming essential for life. Eur. J. Nutr. 2000, 39, 62–66. 10.1007/s003940050003. [DOI] [PubMed] [Google Scholar]
- Kehrer J. P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. 10.1016/S0300-483X(00)00231-6. [DOI] [PubMed] [Google Scholar]
- Rivera M.Bacterioferritin: Structure Function and Protein-Protein Interactions. In Handbook of Porphyrin Science; Kadish K. K., Smith K. M., Guilard R., Eds., 2014; Vol. 30; pp 135–178, DOI: 10.1142/9789814407755_0041 [DOI] [Google Scholar]
- Andrews S. C. The Ferritin-like Superfamily: Evolution of the Biological Iron Storeman from a Ruberythrin-Like Ancestor. Biochim. Biophys. Acta, Gen. Subj. 2010, 1800, 691–705. 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Lundin D.; Poole A. M.; Sjoberg B. M.; Hogbom M. Use of structural phylogenetic networks for classification of the ferritin-like superfamily. J. Biol. Chem. 2012, 287, 20565–20575. 10.1074/jbc.M112.367458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky A. M.; Vakser I. A.; Rivera M. Local packing modulates diversity of iron pathways and cooperative behavior in eukaryotic and prokaryotic ferritins. J. Chem. Phys. 2014, 140, 115104. 10.1063/1.4868229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J. M.; Le Brun N. E.; Moore G. R. Ferritins: furnishing proteins with iron. JBIC, J. Biol. Inorg. Chem. 2016, 21, 13–28. 10.1007/s00775-016-1336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E. C. Ferritin: the protein nanocage and iron biomineral in health and in disease. Inorg. Chem. 2013, 52, 12223–12233. 10.1021/ic400484n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E. C.; Tosha T.; Behera R. K. Solving Biology’s Iron Chemistry Problem with Ferritin Protein Nanocages. Acc. Chem. Res. 2016, 49, 784–791. 10.1021/ar500469e. [DOI] [PubMed] [Google Scholar]
- Honarmand Ebrahimi K.; Hagedoorn P. L.; Hagen W. R. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem. Rev. 2015, 115, 295–326. 10.1021/cr5004908. [DOI] [PubMed] [Google Scholar]
- Frolow F.; Kalb A. J.; Yariv J. Structure of a Unique Twofold Symmetric Haem-Binding Site. Nat. Struct. Biol. 1994, 1, 453–460. 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- Weeratunga S.; Lovell S.; Yao H.; Battaile K. P.; Fischer C. J.; Gee C. E.; Rivera M. Structural Studies of Bacterioferritin B (BfrB) from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center. Biochemistry 2010, 49, 1160–1175. 10.1021/bi9015204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. R.; Kadir H. A.; Al-Massad K.; Le Brun N. E.; Thomson A. J.; Greenwood C.; Keen J. N.; Findlay J. B. C. Structural Heterogeneity of Pseudomonas aeruginosa Bacterioferritin. Biochem. J. 1994, 304, 493–497. 10.1042/bj3040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A.; Wilderman P. J.; Vasil A. I.; Vasil M. L. GeneChip Expression Analysis of the Iron Starvation Response in Pseudomonas Aeruginosa: Identification of Novel Pyoverdine Biosynthesis Genes. Mol. Microbiol. 2002, 45, 1277–1287. 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- Palma M.; Worgall S.; Quadri L. E. N. Transcriptome Analysis of the Pseudomonas aeruginosa Response to Iron. Arch. Microbiol. 2003, 180, 374–379. 10.1007/s00203-003-0602-z. [DOI] [PubMed] [Google Scholar]
- Yao H.; Jepkorir G.; Lovell S.; Nama P. V.; Weeratunga S. K.; Battaile K. P.; Rivera M. Two Disctinct Ferritin-Like Molecules in P. aeruginosa: The Product of the bfrA Gene is a Bacterial Ferritin (FtnA) not a bacterioferritin (Bfr). Biochemistry 2011, 50, 5236–5248. 10.1021/bi2004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.-F.; Ochsner U. A.; Klotz M. G.; Nanayakkara V. K.; Howell M. L.; Johnson Z.; Posey J. E.; Vasil M. L.; Monaco J. J.; Hassett D. J. Bacterioferritin A Modulates Catalase A (KatA) Activity and Resistance to Hydrogen Peroxide in Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 3730–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M. A.; Jordan P.; Grogan J. M.; Butt J. N.; Lutz M.; Thomson A. J.; Andrews S. C.; Guest J. R. Spectroscopic and Voltammetric Characterization of Bacterioferritin-Associated Ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 1996, 229, 635–642. 10.1006/bbrc.1996.1856. [DOI] [PubMed] [Google Scholar]
- Garg R. P.; Vargo C. J.; Cui X.; Kurtz D. M. J. A [2Fe-2S] Protein Encoded by an Open Reading Frame Upstream of the Escherichia coli Bacterioferritin Gene. Biochemistry 1996, 35, 6297–6301. 10.1021/bi9600862. [DOI] [PubMed] [Google Scholar]
- Weeratunga S.; Gee C. E.; Lovell S.; Zeng Y.; Woodin C. L.; Rivera M. Binding of Pseudomonas aeruginosa Apobacterioferritin-Associated Ferredoxin to Bacterioferritin B Promotes Heme Mediation of Electron Delivery and Mobilization of Core Mineral Iron. Biochemistry 2009, 48, 7420–7431. 10.1021/bi900561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Wang Y.; Lovell S.; Kumar R.; Ruvinsky A. M.; Battaile K. P.; Vakser I. A.; Rivera M. The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin. J. Am. Chem. Soc. 2012, 134, 13470–13481. 10.1021/ja305180n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Yao H.; Cheng Y.; Lovell S.; Battaile K. P.; Midaugh C. R.; Rivera M. Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Potein-Protein Interactions in Solution and Determination of Binding Energy Hot Spots. Biochemistry 2015, 54, 6162–6175. 10.1021/acs.biochem.5b00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui H.; Rivera M.; Im W. Protein dynamics and ion traffic in bacterioferritin. Biochemistry 2012, 51, 9900–9910. 10.1021/bi3013388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Rui H.; Kumar R.; Eshelman K.; Lovell S.; Battaile K. P.; Im W.; Rivera M. Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic. Biochemistry 2015, 54, 1611–1627. 10.1021/bi501255r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-L.; Zhou H.-N.; Xing W.-M.; Zhao J.-F.; Li S.-X.; Huang J.-F.; Bi R.-C. 2.6 Å Resolution Crystal Structure of the Bacterioferritin from Azotobacter vinelandii. FEBS Lett. 2004, 573, 93–98. 10.1016/j.febslet.2004.07.054. [DOI] [PubMed] [Google Scholar]
- Swartz L.; Kuchinskas M.; Li H.; Poulos T. L.; Lanzilotta W. N. Redox-Dependent Structural Changes in the Azotobacter vinelandii Bacterioferritin: New Insights into the Ferroxidase and Iron Transport Mechanism. Biochemistry 2006, 45, 4421–4428. 10.1021/bi060146w. [DOI] [PubMed] [Google Scholar]
- Wong S. G.; Grigg J. C.; Le Brun N. E.; Moore G. R.; Murphy M. E.; Mauk A. G. The B-type channel is a major route for iron entry into the ferroxidase center and central cavity of bacterioferritin. J. Biol. Chem. 2015, 290, 3732–3739. 10.1074/jbc.M114.623082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J. M.; Moore G. R.; Le Brun N. E. Mechanisms of iron mineralization in ferritins: one size does not fit all. JBIC, J. Biol. Inorg. Chem. 2014, 19, 775–785. 10.1007/s00775-014-1136-3. [DOI] [PubMed] [Google Scholar]
- Wang A.; Rodríguez J. C.; Han H.; Schönbrunn E.; Rivera M. X-Ray Crystallographic and Solution State Nuclear Magnetic Resonance Spectroscopic Investigations of NADP+ Binding to Ferredoxin NADP Reductase from Pseudomonas aeruginosa. Biochemistry 2008, 47, 8080–8093. 10.1021/bi8007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.; Zeng Y.; Han H.; Weeratunga S.; Morgan B. N.; Moënne-Loccoz P.; Schönbrunn E.; Rivera M. Biochemical and Structural Characterization of Pseudomonas aeruginosa Bfd and FPR: Ferredoxin NADP+ Reductase and Not Ferredoxin is the Redox Partner of Heme Oxygenase under Iron-Starvation Conditions. Biochemistry 2007, 46, 12198–12211. 10.1021/bi7013135. [DOI] [PubMed] [Google Scholar]
- Richards T. D.; Pitts K. R.; Watt G. D. A Kinetic Study of Iron Release from Azotobacter Vinelandii Bacterial Ferritin. J. Inorg. Biochem. 1996, 61, 1–13. 10.1016/0162-0134(95)00029-1. [DOI] [PubMed] [Google Scholar]
- Yasmin S.; Andrews S. C.; Moore G. R.; Le Brun N. E. A New Role for Heme, Facilitating Release of Iron from the Bacterioferritin Iron Biomineral. J. Biol. Chem. 2011, 286, 3473–3483. 10.1074/jbc.M110.175034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O.; Gursoy A.; Ma B.; Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact?. Chem. Rev. 2008, 108, 1225–1244. 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]