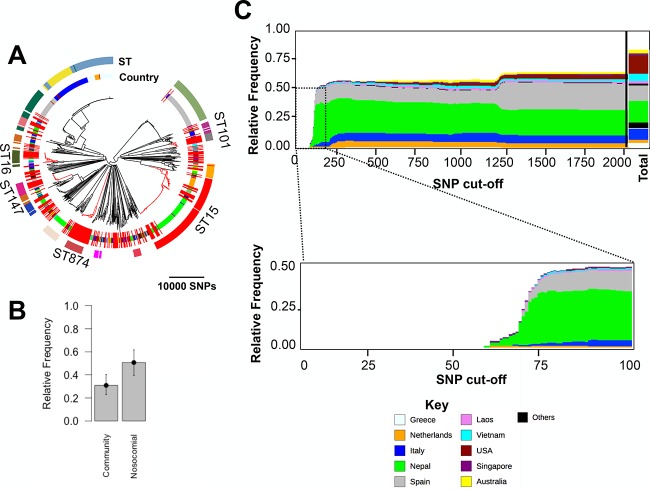
FIG 2 .
(A) A neighbor-joining phylogenetic tree for the KpI lineage depicting the linkages between isolates in the United Kingdom and Ireland collection and those in the global collection with ≤10,000 SNPs divergence from them (equivalent to membership in the same ST). The outer circular band shows STs that were represented by at least 10 isolates, and the next band inward shows the country of origin according to the key in Fig. 1. The clades in red are the major STs that contained at least 10 United Kingdom and Ireland MDR isolates. (B) The relative frequencies of global isolates in the phylogenetic tree in panel A based on the source of acquisition of infection for those isolates in the global study of Holt et al. (4). The error bars denote 95% confidence intervals obtained from the binary distribution. (C) The origin of the closest non-United-Kingdom relatives of United Kingdom and Ireland isolates in the global population shown at increasing SNP cutoffs from left to right. The total column represents the proportion of isolates in the combined tree. United Kingdom and Ireland MDR K. pneumoniae isolates are not shown.