Extended Data Figure 9. Cell-type specificity and reproducibility of Hi-C interactions with schizophrenia GWAS hits.
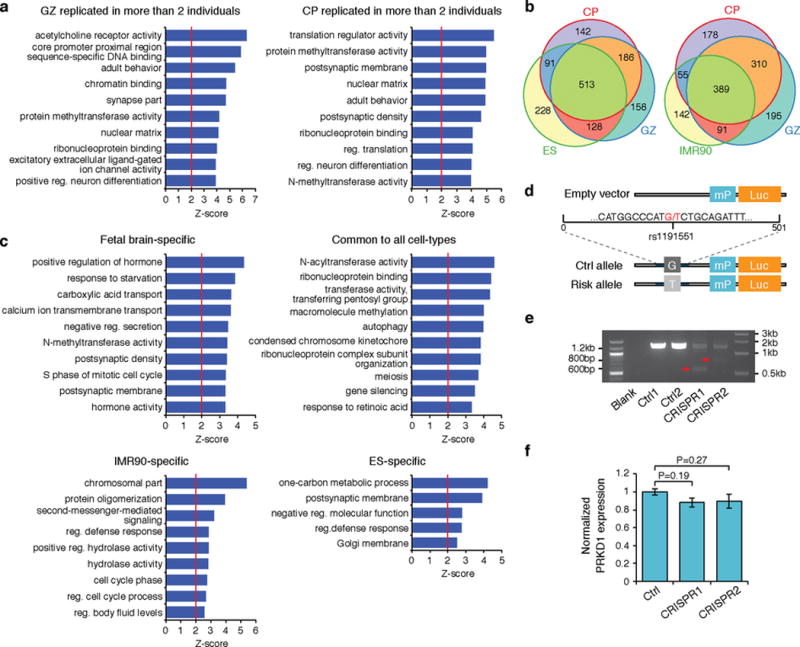
a, GO enrichment for schizophrenia risk genes replicated in more than two individuals in CP (left) and GZ (right). b, Overlap between genes that interact to schizophrenia credible SNPs in CP and GZ vs ES (left) and IMR90 (right). c, GO enrichment for genes that interact to schizophrenia credible SNPs in cell-type specific manner. d, Schematic showing the incorporation of sequence flanking rs1191551 into reporter (Luc) vector with a minimal promoter (mP). e, PCR amplification of targeted genomic region demonstrates deletion of the SNP-containing region. Expected band size, 587 bp (CRISPR1) and 813 bp (CRISPR2). f, CRISPR/Cas9-mediated deletion of rs1191551 flanking region does not affect the closest protein-coding gene PRKD1 expression. Normalized expression levels of PRKD1 relative to control (Ctrl) (mean ± standard error, n = 6 (Ctrl), 4 (CRISPR1 and CRISPR2)). P values, one-way ANOVA and post hoc Tukey test.
