Figure 1. Sil1 regulates BiP oxidation state in cells.
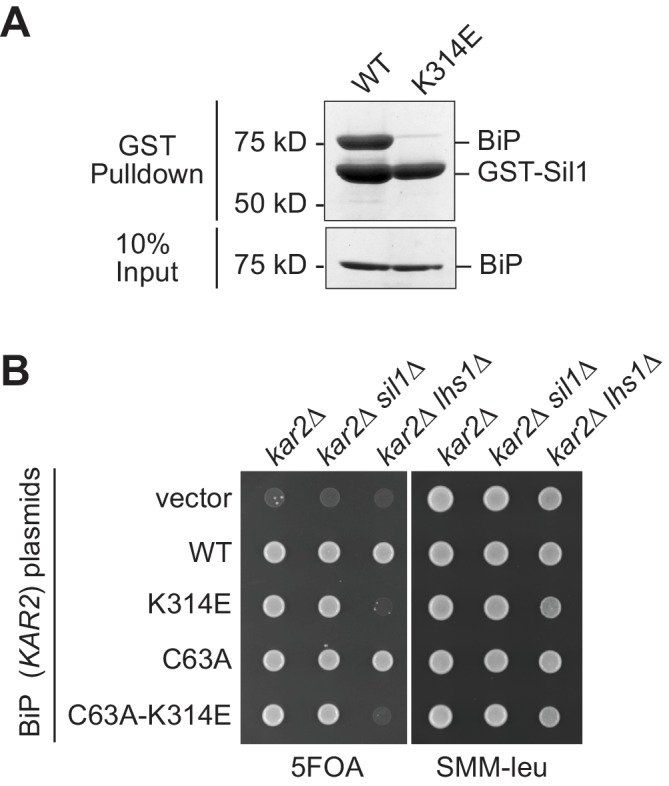
(A) Yeast strains (CSY289, 290, 612, 689) were spotted onto SMM plates containing 0–1.5 mM diamide and incubated for 2 d at 30°C. (B) Schematic for the biotin-switch procedure. (C) Yeast strains deleted for endogenous BiP (kar2∆) containing plasmids encoding FLAG-tagged BiP were assayed for oxidized BiP levels using the biotin-switch protocol. Oxidative stress was generated by overexpression of Ero1*. BiP was immunoprecipitated, and total and oxidized BiP were detected by Western blotting. The relative levels of oxidized BiP are expressed as the ratio of the intensity of the avidin and anti-BiP signals. The signal ratio was set to 1.0 for wild-type cells grown in the absence of Ero1*. (D) Lysates were prepared from the indicated yeast after Ero1* induction. Oxidized BiP levels were detected and quantified as in C. (E) Yeast strains (CSY5, 275, 448, 449) were spotted onto YPD plates containing 0–2.0 mM diamide and were incubated for 2 d at 30°C. (F) Cells were treated with 5 mM diamide for 15 min, diamide was removed, and cells were returned to 30°C until harvest. Oxidized BiP levels were determined as in C. (G) Plot of the averaged quantified data ± SEM from F and a second independent experiment using the same protocol. For each strain, the signal ratio was set to 1.0 for cells grown without diamide.
Figure 1—figure supplement 1. BiP-Sil1 structure.
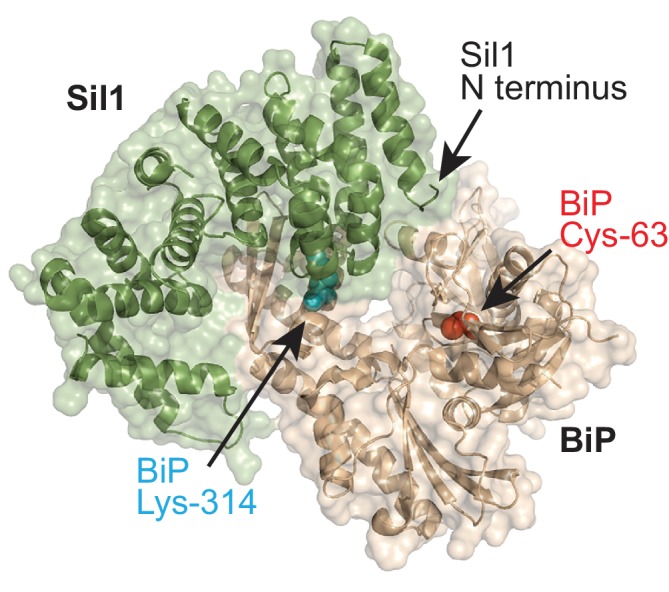
Figure 1—figure supplement 2. A BiP K314E mutation disrupts Sil1 binding.