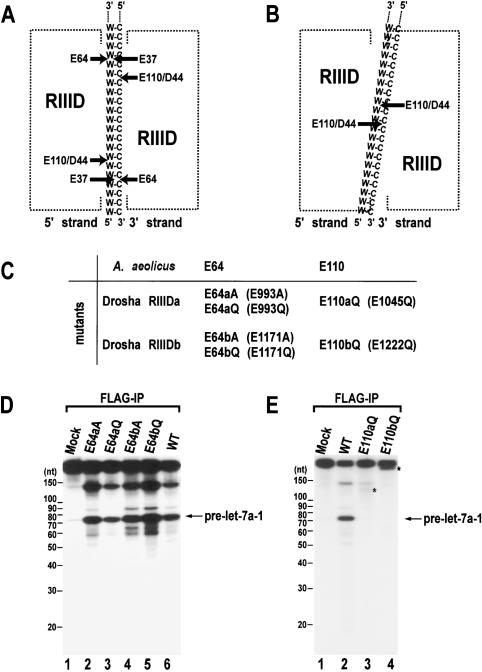
Figure 1.
Site-directed mutagenesis of human Drosha. (A) A model for the cleavage mechanism of RNase III (Blaszczyk et al. 2001). Two processing centers are formed between two RIIIDs. Each center contains two catalytic sites that cleave two nearby phosphodiester bonds on the opposite RNA strands of dsRNA (W-C, Watson-Crick base pairs). The 5′ strand indicates the strand that contains the 5′ phosphate group at the terminus of stem. The 3′ strand means the strand that possesses the 3′-hydroxyl group at the 3′ end. (B) The “single processing center” model where only two catalytic sites are formed in a single processing center (Zhang et al. 2004). (C) Naming of the single amino acid mutants in the RIIIDs of human Drosha. (D) In vitro processing of pri-let-7a-1. The substrate was incubated with E64aA, E64aQ, E64bA, E64bQ, or wild-type (WT) Drosha-Flag proteins that were expressed in HEK293T cells and prepared by immunoprecipitation with anti-Flag antibody. The RNA size markers (Decade Marker, Ambion) are indicated on the left side of the gel. (E) In vitro processing of pri-let-7a-1. The substrate was incubated with E110aQ, E110bQ, or wild-type (WT) Drosha-Flag proteins that were immobilized on anti-Flag beads. The asterisks indicate fragments that accumulate when the particular mutant was used in the assay.