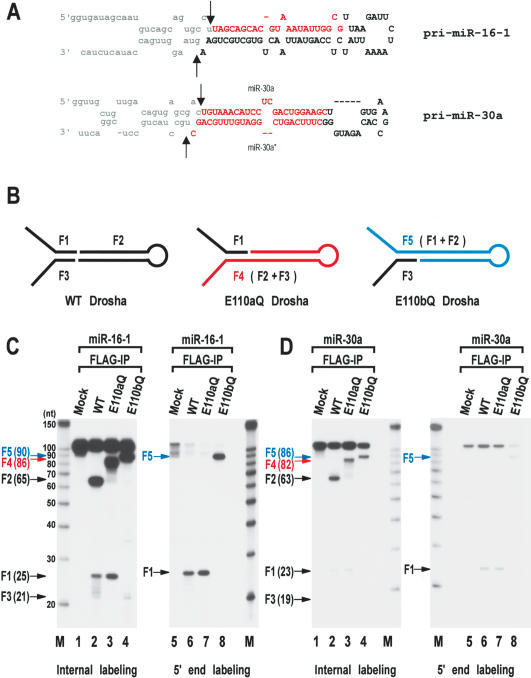
Figure 2.
Processing of the “minimal” pri-miRNA with E110 mutants. (A) Sequences and secondary structure of the short pri-miRNAs derived from pri-miR-16-1 and pri-miR-30a. These RNAs are 111 nt and 105 nt, respectively. The cleavage sites (the 5′ and 3′ ends of pre-miRNAs) are indicated with arrows. Actual cleavage sites of pri-miR-16-1 were determined by directional cloning of pre-miR-16-1. Pre-miR-16-1 was prepared by in vitro processing, ligated to the 5′ and 3′ adapters, amplified by RT-PCR, and subcloned into pGEM-T-easy vector. Ten clones were randomly chosen and sequenced. All the clones were identical at the 5′ end. Five of the 10 clones had the same 3′ end shown here. Three other clones were 1 nt shorter and two other clones were 2 nt shorter at the 3′ end, which is likely to be due to exonucleolytic trimming. The cleavage sites at miR-30a were reported (Lee et al. 2003). Pre-miR-16-1 shown here is 65 nt; pre-miR-30a is 63 nt. (B) Schematic representation of the processing products from in vitro processing by wild-type (WT), E110aQ, and E110bQ Drosha. Wild-type Drosha generates F1, F2, and F3 E110aQ produces F1 and F4 and E110bQ generates F3 and F5. (C) In vitro processing of pri-miR-16-1. The short pri-miR-16-1 was incubated with wild-type (WT) Drosha, E110aQ, or E110bQ proteins that were expressed in HEK293T cells and immobilized on anti-Flag affinity gel. The substrate was either internally labeled during transcription (lanes 1-4) or labeled at the 5′ end after transcription (lanes 5-8). The size of each fragment is indicated in the bracket next to the arrow. The RNA size markers (Ambion) are indicated on the side of the gel. (D)In vitro processing of pri-miR-30a. The short pri-miR-30a was incubated with wild-type Drosha, E110aQ, or E110bQ proteins as in C. The substrate was either internally labeled during transcription (lanes 1-4) or labeled at the 5′ end after transcription (lanes 5-8).