Figure 3.
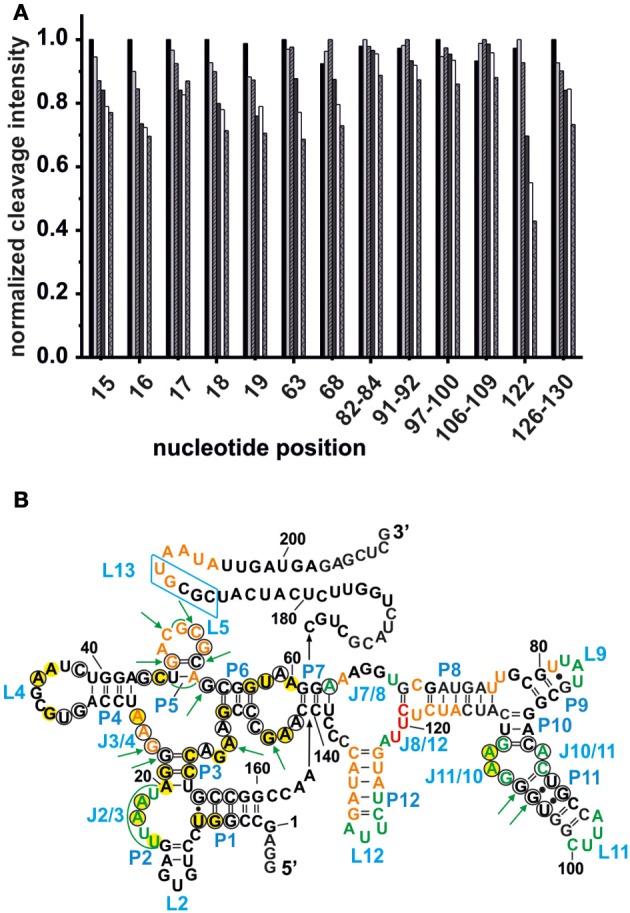
Competition between Mg2+ and Tb3+ for the identification of specific Mn+ binding sites. (A) The decrease in cleavage intensity at the mapped sites is shown as a function of Mg2+ while Tb3+ was held constant at 500 μM. The Mg2+ concentration increases from 20 to 90 mM (from left-to-right). (B) Comparison of the metal ion binding sites of the AdoCbl riboswitches from E. coli and S. thermophilum (Peselis and Serganov, 2012). Shown is the proposed secondary structure of the btuB riboswitch with circled nucleotides representing the conserved bases in the consensus sequence of AdoCbl riboswitches (Nahvi et al., 2004). Green nucleotides correspond to specific Mg2+ binding sites of the btuB riboswitch. The nucleotides marked by green arrows indicate the metal ion binding sites at conserved nucleotides as found in the S. thermophilum AdoCbl riboswitch while areas marked in green signal possible metal ion binding sites at non-conserved nucleotides. Bases marked in yellow could be involved in tertiary interactions as comparison with the S. thermophilum riboswitch suggests (Peselis and Serganov, 2012). The blue box indicates the L13 region supposedly undergoing the kissing loop interaction with L5 upon AdoCbl binding. This tertiary interaction is not visible in the crystallized riboswitch from S. thermophilum since it was truncated after P1. Orange and red nucleotides were shown to provide weak and strong Tb(III) binding sites respectively (see Figure 2).
