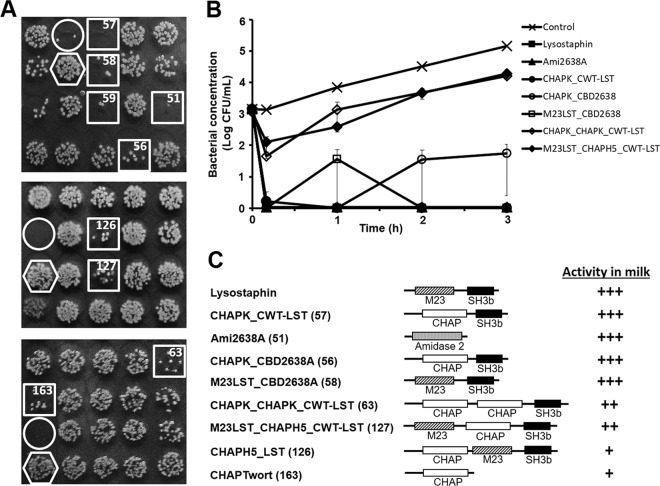
FIG 1.
Identification of PGH constructs with high activity against S. aureus in milk. (A) Three sections from screening plates used for identification of promising PGH constructs (see the text for details). E. coli strains harboring different PGH constructs were grown in 96-well plates. Following protein production, cells were harvested and enzymes were liberated by exposure of pellets to chloroform vapor. UHT milk spiked with S. aureus Newbould 305 was added to the wells and incubated for 2 h before aliquots of each well were spotted on agar plates. Spots with largely reduced S. aureus colony density after overnight incubation (squares) suggest the presence of promising PGH constructs in the corresponding wells. A lysostaphin-expressing E. coli strain (circles) and a strain harboring no PGH construct (hexagons) served as positive and negative controls, respectively. Numbers in the squares correspond to PGH constructs shown in panel C. (B) Comparative time-kill assay with S. aureus Newbould 305 cells in UHT milk using purified PGH constructs (360 nM) that had been identified as promising candidates by the screening as exemplified in panel A or buffer as control. Bacterial concentrations at different time points after enzyme/buffer addition were determined by serial dilution plating. It should be noted that symbols for lysostaphin, Ami2638A, and CHAPK_CWT-LST appear on top of each other since these constructs caused instantaneous reduction of S. aureus to undetectable levels. Error bars represent standard errors of the means from 3 experiments. (C) Schematics of candidate PGH constructs rated for their activity in milk based on log reduction of S. aureus cells compared to the control after 3 h in time-kill assays as exemplified in panel B. Construct numbers are given in parentheses. Note that construct 59 (A) is an N-terminally 6×His-tagged version of the positive control lysostaphin and was therefore not included in the ranking. +, <3 log reduction at 4 μM; ++, ≥3 log reduction at 4 μM but <3 log reduction at 360 nM; +++, ≥3 log reduction at 360 nM. M23, M23 endopeptidase domain; CHAP, cysteine, histidine-dependent amidohydrolase/peptidase domain; Amidase 2, amidase 2 domain; SH3b, bacterial SH3 cell wall binding domain.