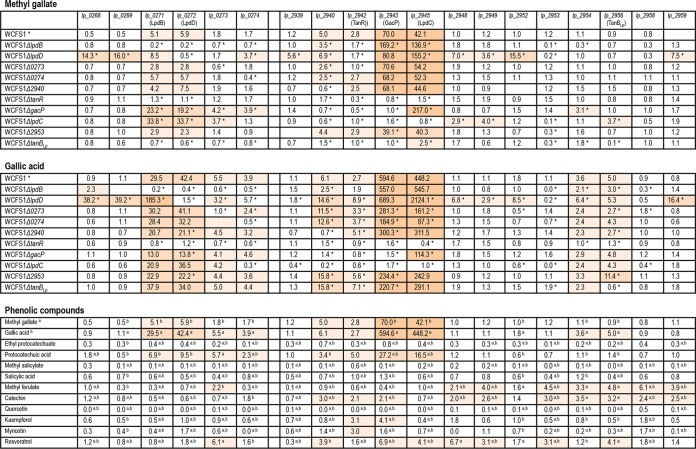
FIG 4.
Relative transcriptional expression of genes putatively related to tannin metabolism in L. plantarum WCFS1 inoculated onto a medium containing methyl gallate, gallic acid, or one of several phenolic compounds. These compounds were added at 3 mM (for methyl gallate, gallic acid, ethyl protocatechuate, protocatechuic acid, methyl salicylate, salicylic acid, methyl ferulate, and catechin), 0.3 mM (quercetin), 10 μM (kaempferol and myricetin), and 1 mM (resveratrol) concentrations. Relative expression on methyl gallate and gallic acid was assayed for the L. plantarum WCFS1 wild-type strain and for 10 mutants with knockouts of genes putatively involved in tannin metabolism. RQ values were calculated by the comparative CT method using the 7500 Fast System relative quantification software with the L. plantarum ldh gene as the endogenous gene and growth in the absence of added phenolic compounds as the growth condition calibrator. The experiments were carried out in triplicate, and the mean values are shown. Uncolored boxes show RQ values lower than 2 (an RQ value of 2 indicates a 2-fold increase in gene expression over an RQ value of 1, which is assigned to expression in the absence of phenolic compounds). Light-, medium- and dark-pink boxes show 2- to 10-fold, 10- to 50-fold, and >50-fold increases in RQ values, respectively, over those in the absence of phenolic compounds. Asterisks indicate significant differences between the wild-type strain and the mutant strain in response to the presence of methyl gallate or gallic acid (P, ≤0.05 by Duncan's test). Superscript letters indicate significant differences between the presence of methyl gallate (a) or gallic acid (b) and the presence of other phenolic compounds (P, ≤0.05 by Duncan's test).