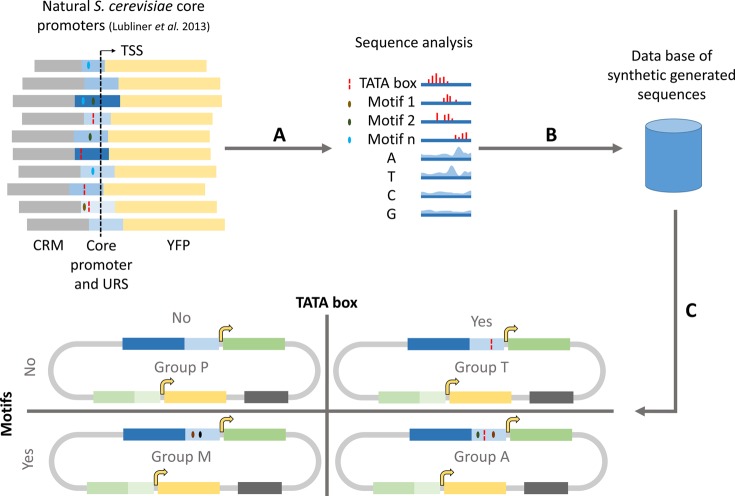
Figure 1.
Design strategy for synthetic core promoters. Three steps were followed: (A) Computation of (i) nucleotide probability distribution, (ii) TATA box position, (iii) position and frequency distribution of motifs and (iv) average nucleosome occupancy along the sequence of 140 S. cerevisiae natural strong core promoters28 aligned by the transcription start site. (B) Generation of 400 random sequences using information on nucleotide probability distribution. (C) Partitioning of sequences in four groups (dubbed P, T, M and A), to which TATA boxes and motifs were added, according to the group they belonged to (group P: without TATA box nor motifs; group T: with TATA box and without motifs; group M: with motifs and without TATA box; group A: with TATA box and motifs). Subsequently, the nucleosome occupancy profile of each of the generated sequences was compared to the average profile for the natural strong promoters. The generated sequences with higher similarity to the average natural nucleosome occupancy were selected to be tested in vivo.