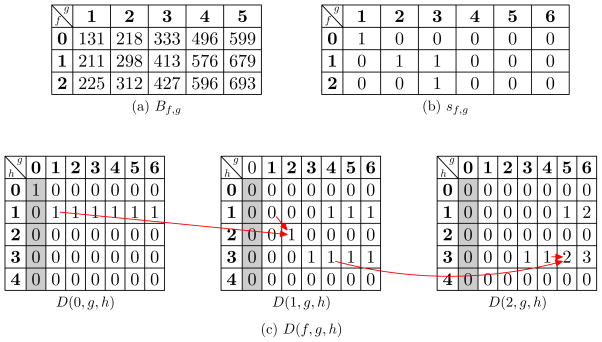
Figure 2.
The three-dimensional table D(f, g, h) for a discretized spectrum with a precursor mass 848 and four neutral fragment masses 131, 413, 421, 550, a protein sequence MSDYCH, and an ordered pair of modifications (phosphorylation, methylation). A scale factor 1 is used in the computation. (a) B0,g is the sum of the masses of the first g residues of the protein. B1,g is the sum of B0,g and the mass of phosphorylation (80 Da). B2,g is the sum of B0,g and the masses of phosphorylation (80 Da) and methylation (14 Da). (b) Table sf,g is generated based on Bf,g using Equation (3). (c) D(f, g, h) is filled out by the dynamic programming algorithm in Figure S1 in the supplementary material. The shaded areas are initialized using Equation (4). The second residue S is a modification site of phosphorylation, and the value D(1, 2, 2) is computed as D(0, 1, 2−s1,2+D(1, 1, 2−s1,2) = D(0, 1, 1)+D(1, 1, 1). Similarly, the fifth residue C is modification site of methylation, and the value D(2, 5, 3) is computed as D(1, 4, 3 − s2,5) + D(2, 4, 3 − s2,5) = D(1, 4, 3) + D(2, 4, 3).