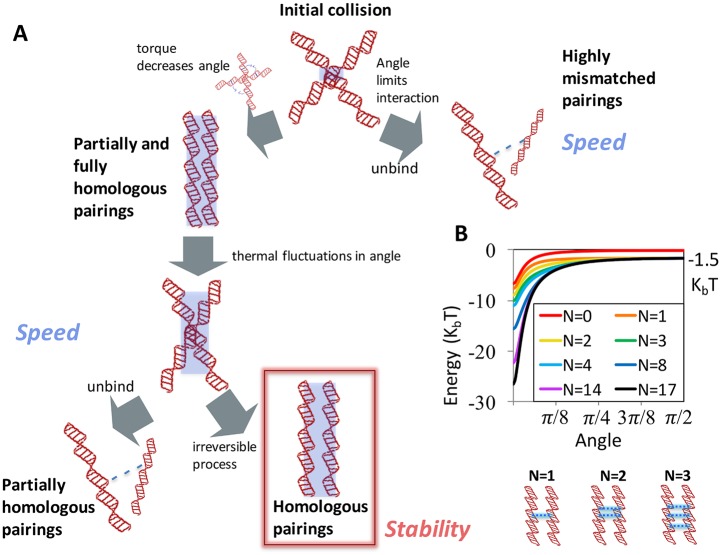
Fig 1. Model for interactions between dsDNA molecules.
(A) Schematic of our model for multi-stage kinetic proofreading. Two DNA molecules (modeled as rigid rods) initially collide at a finite angle. The angle limits contact between interaction sites and ensures that highly mismatched pairings rapidly unbind irreversibly. Meanwhile, pairings with roughly 4 or more matching sites flanking the collision site experience an attractive torque that draws them into a more deeply bound, parallel state. Partially homologous pairings that reach this deeply bound state are destabilized by diffusive fluctuations in angle, which allow pairings to re explore larger angles, from which point irreversible unbinding is faster. These fluctuations provide further speed by allowing a given stringency to be attained faster. Finally, pairings that remain bound for a minimum amount of time are irreversibly stabilized, ensuring stability for homologs. In this diagram, the blue shading indicates the approximate length over which pairings interact. (B) Energy as a function of angle for pairs of rods with a variable number of continuous matched sites flanking the collision point, N. Example diagrams of pairings with N = 1, 2, and 3 are shown below the plot, with the blue, dashed lines indicating attractive interactions between matched pairs of sites. The attractive energy per matched site is -1.5 kB T, marked on the plot.