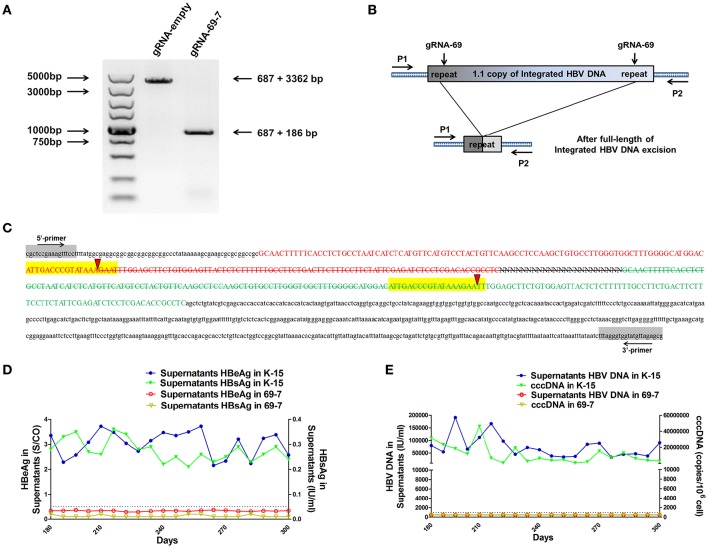
Figure 3.
CRISPR-Cas9/gRNA-69 efficiently removed the integrated HBV genome from a stable HBV cell line. (A) Analysis of PCR amplicon lengths using a primer pair (P1 and P2) targeting the integrated HBV-flanking sequence revealed elimination of the full-length integrated HBV genome (3,173-bp), leaving one fragment (873-bp predicted segment from its flanking region). (B) Diagram showing excision of the full-length integrated HBV genome. The remaining fragment included the expected 687-bp from the integrated HBV flanking sequence and a 186-bp HBV repeat core region sequence. (C) Sanger sequencing of the remaining fragment (873-bp) showing the HBV flanking sequence (small letters, 687-bp) and the partial sequences (189 − 3 = 186-bp) of the integrated HBV repeat region B (green) and repeat region A (red) with a 3-bp deletion around the gRNA-69 targeting site (yellow-highlighted). Elimination of the full-length integrated HBV genome is indicated by a strikethrough. (D,E) The amounts of HBeAg, HBsAg and HBV DNA in cell culture supernatants and HBV cccDNA in the gRNA-empty-treated group (K-15) and gRNA-69-treated group (69-7) over 300 consecutive days. The HBsAg and HBeAg test results in the gRNA-69-treated group (69-7) were always under the negative threshold (0.05 IU/ml for HBsAg and 1 S/CO for HBeAg), and the amounts of HBV DNA and HBV cccDNA in the supernatants were always undetectable (<500 IU/ml for qPCR) in the gRNA-69-treated group (69-7). All viral markers in the gRNA-empty-treated group (K-15) remained at high levels.