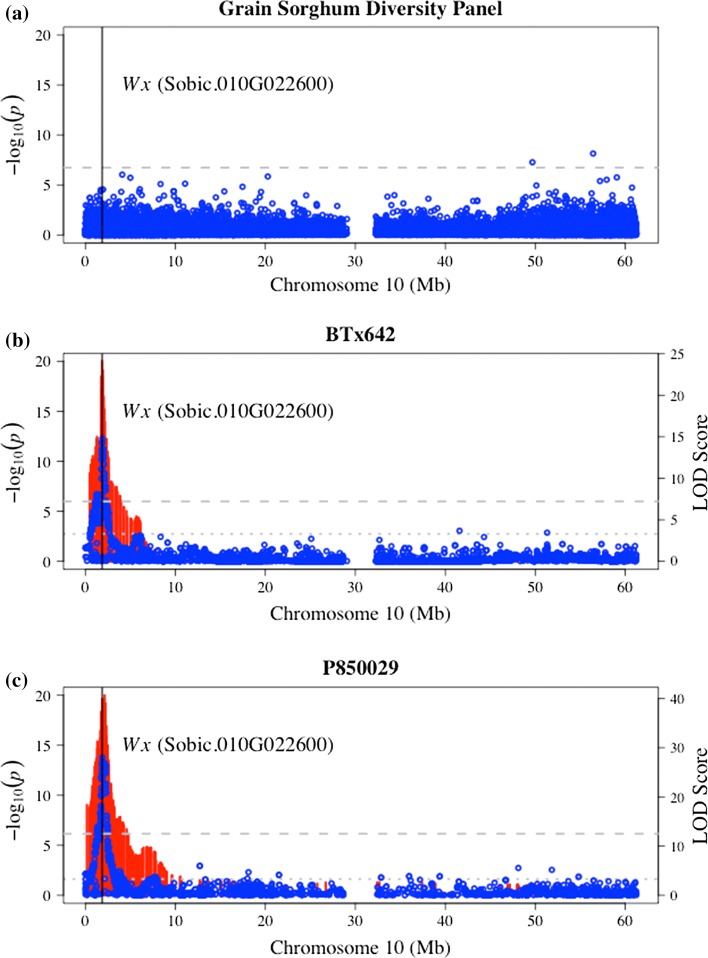
Fig. 3.
Association mapping of amylose across the grain sorghum diversity panel (GSDP) and the two RIL populations segregating for the waxy trait (low amylose %). a As a result of few waxy genotypes in the GSDP and thus a very low minor allele frequency (MAF), no significant associations surrounding the waxy (Wx) locus (black vertical line) are detected. Strong association peaks at the Wx locus are detected using phenotypic data from b BTx642 and c P850029. GAPIT software (Lipka et al. 2012) using the full SNP data set (blue circles) and R/qtl software (Broman et al. 2003) using recombination bin markers (red lines) both easily identified Wx at high resolution. The SNP with highest average significance between the two RIL populations was located 12 kb from Wx (Sobic.010G022600)