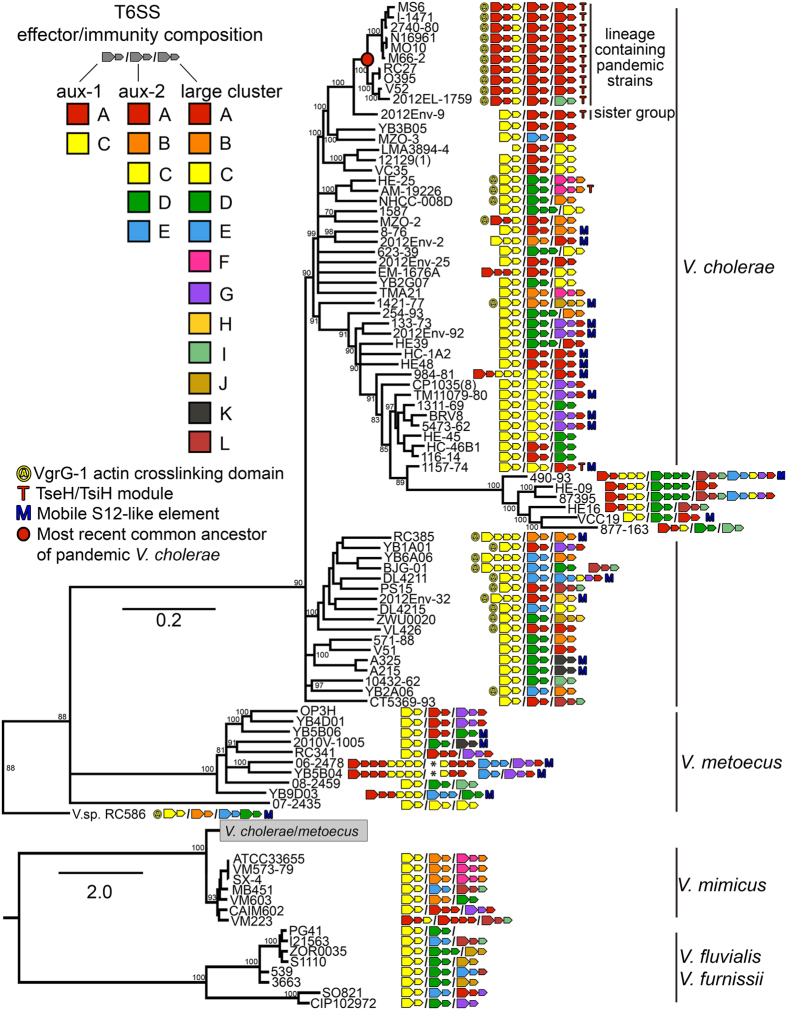
Figure 2. Whole-genome phylogeny and T6SS EI module composition of Vibrio cholerae and closely related species.
Large arrows next to strain names indicate effector, small arrows immunity genes. Auxiliary cluster 1, 2 and the large cluster are separated by slashes. Asterisks indicate transposons. The phylogenetic tree was calculated using the GTR + Gamma Maximum likelihood model implemented in RAxML based on a 1,085,207 pangenome 19-mer alignment (including not just characters shared by all, but by at least two genomes) created using kSNP3. V. cholerae/V. metoecus are visualized separately for better visibility of short internal branches. Statistical branch support was obtained from 100 bootstrap repeats. Bootstrap support for relevant bipartitions is indicated, and branches with support <70 were collapsed. Scale bar indicates substitutions/site. Accession numbers of genomes are listed in Supplementary Table 1.