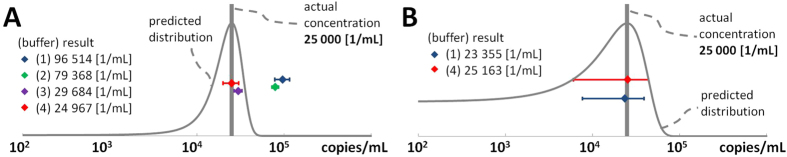
Figure 5. Experimental verification of a 16-compartment digital assay with comparison to the performance of qPCR assays.
(A) The graph shows the results for the same amount of reference DNA suspended in different elution buffers and quantified with conventional qPCR and with the synergistic PCR algorithm. Tests performed on Applied Biosystems 7500 Fast RT System on the IVD Cytomegalovirus PCR kit (GeneProof) according to the prescription. Elution buffers: (1) water, (2) AE elution buffer QIAamp DNA Mini Kit (Quiagen), (3) MBL5 NucleoMag Blood 200 uL (MACHEREY-NAGEL), (4) MagJET Whole Blood Genomic DNA Kit (Thermo Scientific). The gray line shows the expected distribution of results for Real-Time assay. (B) The graph shows the result of 24 runs of the synergistic assay, each on 16 partitions of the amplification mix. We conducted two series of 12 assays on the two elution buffers (1 and 4) that provided the largest difference in the result of the conventional qPCR analysis. The gray line shows the expected distribution of results from the synergistic assay used in the experiment, which should provide 60% precision of assessment. This distribution was also verified using 10,000 Monte-Carlo simulations.