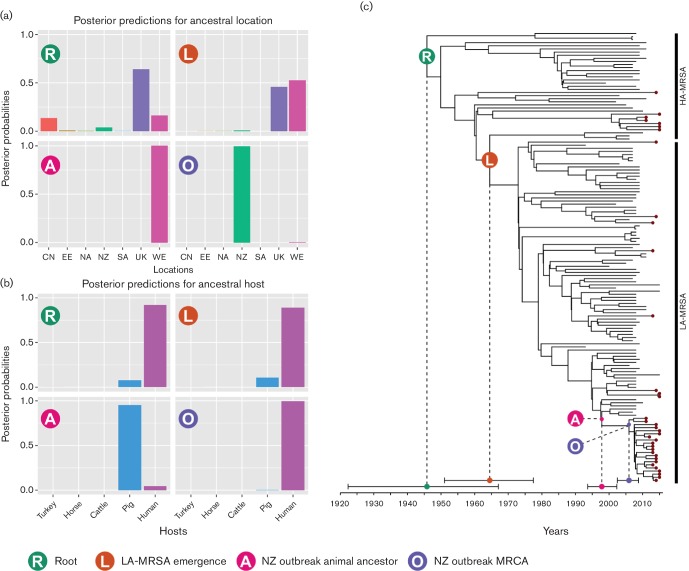
Fig. 3.
Bayesian evolutionary analysis demonstrating the timing of divergence within the CC398 clade, as well as the most likely location and host for a subset of four relevant nodes: R, the MRCA of all CC398 samples; L, the MRCA of LA-MRSA; A, the non-human ancestor of the most recent NZ LA-MRSA outbreak; O, the human MRCA of the most recent NZ LA-MRSA outbreak. (a) Posterior probability mass distribution of geographical locations for four highlighted nodes. (b) Posterior probability mass distribution of host species for four highlighted nodes. (c) Maximum clade credibility genealogy for CC398 samples, depicting the median and 95 % HPD for age of the four important nodes. Isolates from NZ are labelled with a burgundy tip. NZ, New Zealand; SA, South America; EE, Eastern Europe; WE, Western Europe; CN, China; UK, United Kingdom; HA-MRSA, human-associated MRSA.