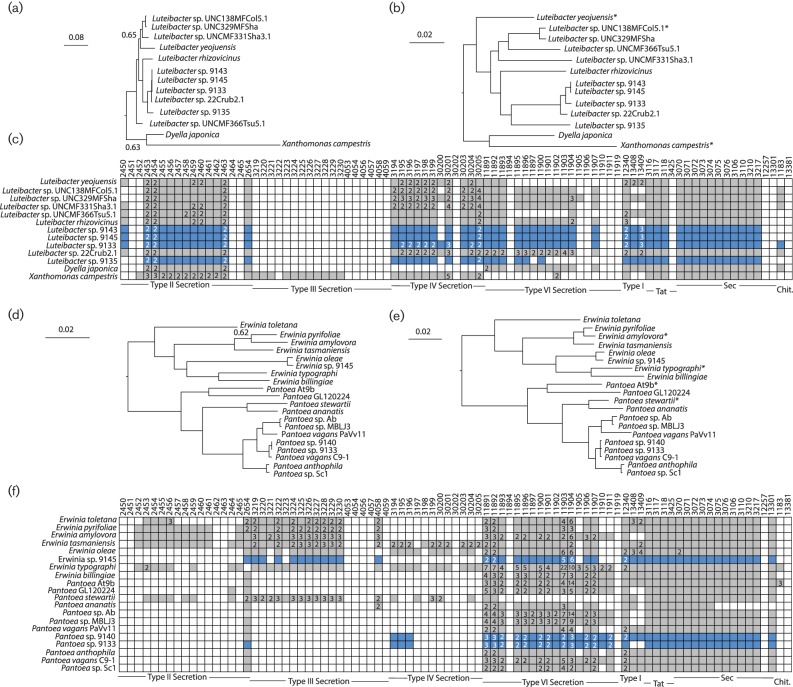
Fig. 1.
Phylogenetic analysis and comparison of interdomain interaction systems for Erwinia, Pantoea and Luteibacter. (a, d) Bayesian phylogenies for focal EHB and non-EHB strains for Leutibacter (a) or Erwinia/Pantoea (d) were inferred from concatenated sequences of RpoD and GyrB. Unless noted, posterior probabilities at all nodes are >0.95. (b, e) Maximum-likelihood phylogenies for EHB and non-EHB strains were inferred from whole-genome sequences using RealPhy for Leutibacter (b) or Erwinia/Pantoea (e). (c, f) KEGG pathway searches were implemented in IMG to identify bacterial pathways known to be involved in signalling between bacteria and eukaryotes for Leutibacter (c) or Erwinia/Pantoea (f). Genomes queried for each clade are listed across the y-axis. Boxes along the x-axis indicate KEGG pathway identifiers (top) for constituent genes for each bacteria secretion system with grouping by system (bottom). Coloured/filled boxes indicate that at least one gene within the genome is present and classified according to that specific KEGG identifier. Numbers inside the coloured/filled boxes denote that more than one gene within that genome is classified according to that KEGG identifier. The boxes for EHB bacteria described in this report are coloured blue. *Indicates that these genomes were used as references for building phylogenies using RealPhy.