Fig. 5.
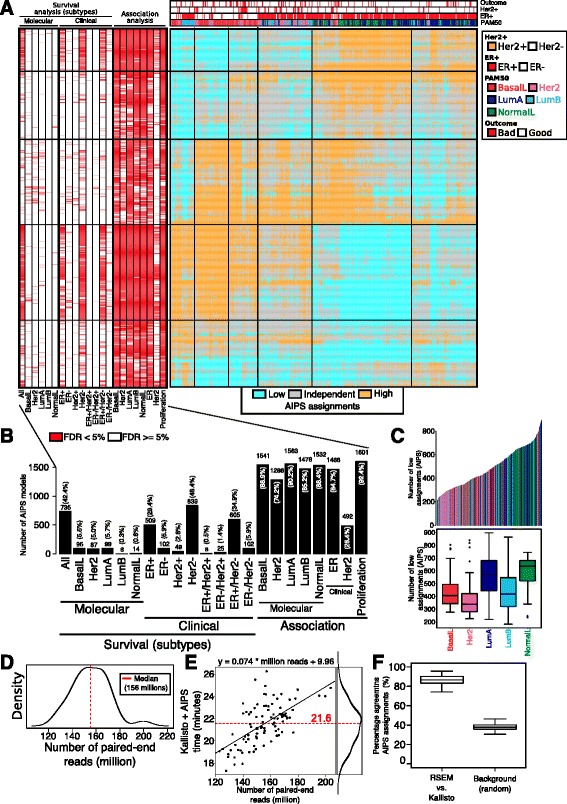
Absolute inference of patient signatures (AIPS) models are prognostic. a Right heatmap depicts the global partitions made by AIPS on the McGill validation dataset (n = 429, cyan low, gray independent, orange high). Left significant associations (false discovery rate (FDR) <5%) induced from AIPS partitions. Far left associations with survival (log-rank test) using the molecular and clinical subtypes and far right significant overlap with the clinical and proliferation molecular subtypes (Fisher’s exact test). b Presentation of the numbers and percentages of AIPS partitions that are significantly (FDR <5%) associated with either survival or overlap. c Top each bar represents the number of times a patient in the McGill validation dataset was assigned to the low class across all 1733 AIPS partitions. Color indicates the subtype of the patient. Bottom distribution of number of times patients of each subtype were assigned to the low class using AIPS partitions. d Distribution of the number of 100-bp paired-end reads use in the Kallisto quantification (n = 96). e Time taken in minutes for Kallisto quantification and AIPS partitions as a function of the number of paired-end reads. Line represents a linear fit of the data. f Kallisto versus RSEM AIPS partitions agreement versus a background distribution agreement obtained from randomly shuffling the labels 100 times. BasalL, basal-like, HER2 human epidermal growth factor receptor 2, LumA luminal A, LumB luminal B, ER estrogen receptor, NormalL normal-like
