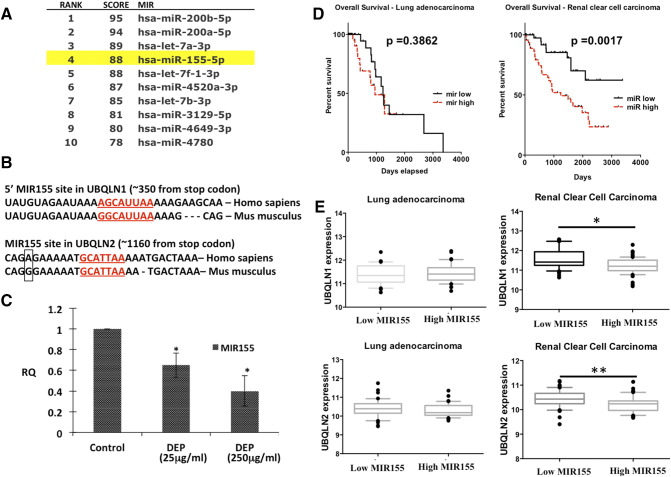
Figure 2.
MIR155 is down-regulated by DEP and is predicted to bind and regulate UBQLN1 and UBQLN2 mRNA. A) Ranking of UBQLN1 targeting miRNAs using miRDB web portal according to their functional annotations (higher the score more the probability of binding); B) Targeting site of MIR155 in 3′-UTR of UBQLN1 and UBQLN2; C) Effect of DEP exposure on expression of MIR155 in A549 cells; D) Kaplan–Meier plot of overall survival of lung adenocarcinoma patients (top) and renal cell carcinoma patients (bottom) stratified by high (dashed) and low (solid) MIR155 expression. E) Box and whiskers plots comparing RSEM of UBQLN expression values in samples with high MIR155 expression (90th percentile) to samples with low MIR155 expression (10th percentile). The box extends from the 25th to the 75th percentile, with the median shown, whereas the whiskers range from 10th to 90th percentile. (Mann–Whitney test, one asterisk P < .05, two asterisks P < .01). Cancer types with a negative correlation between MIR155 and UBQLN expression also had MIR155 expression profiles significantly associated with overall patient survival.