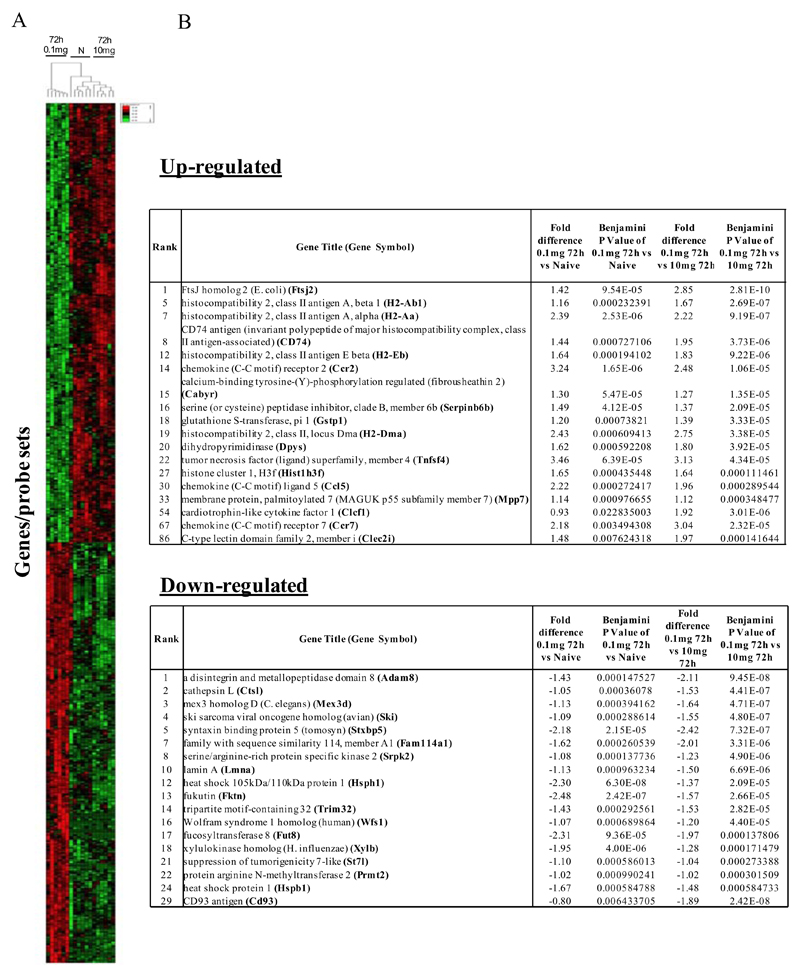
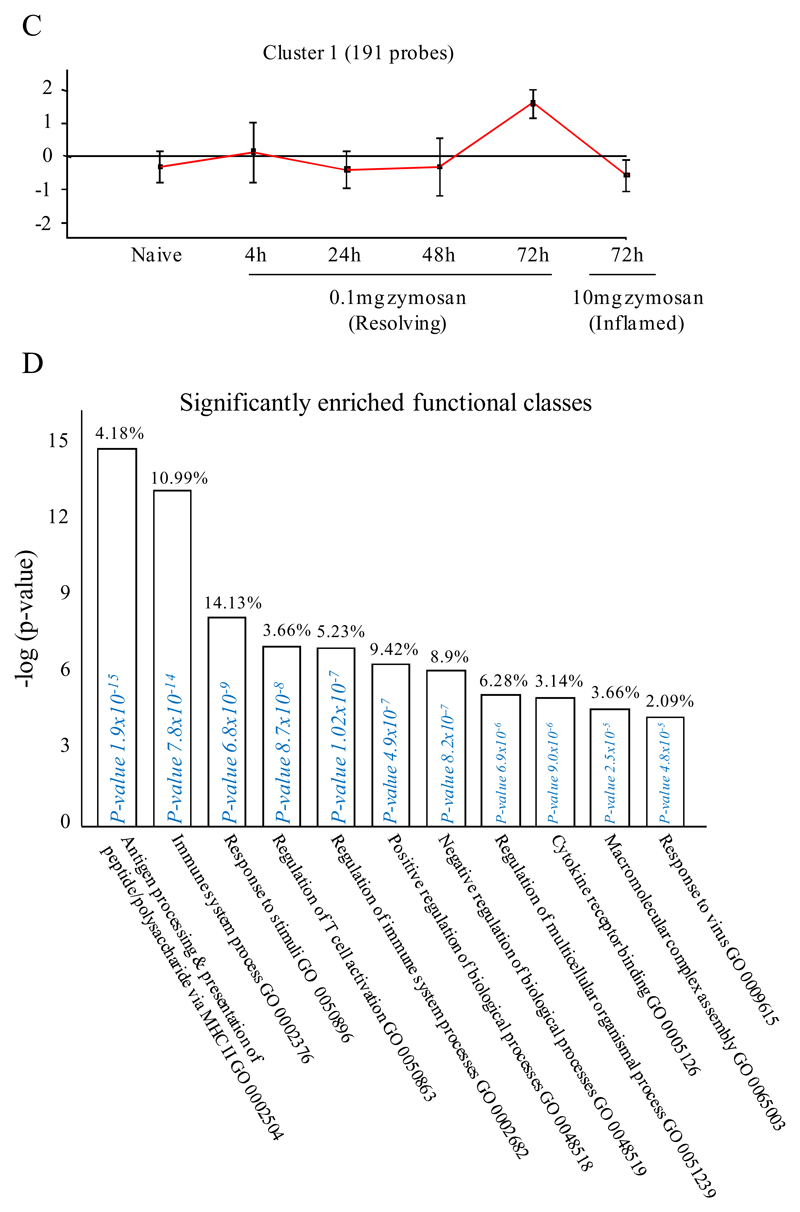
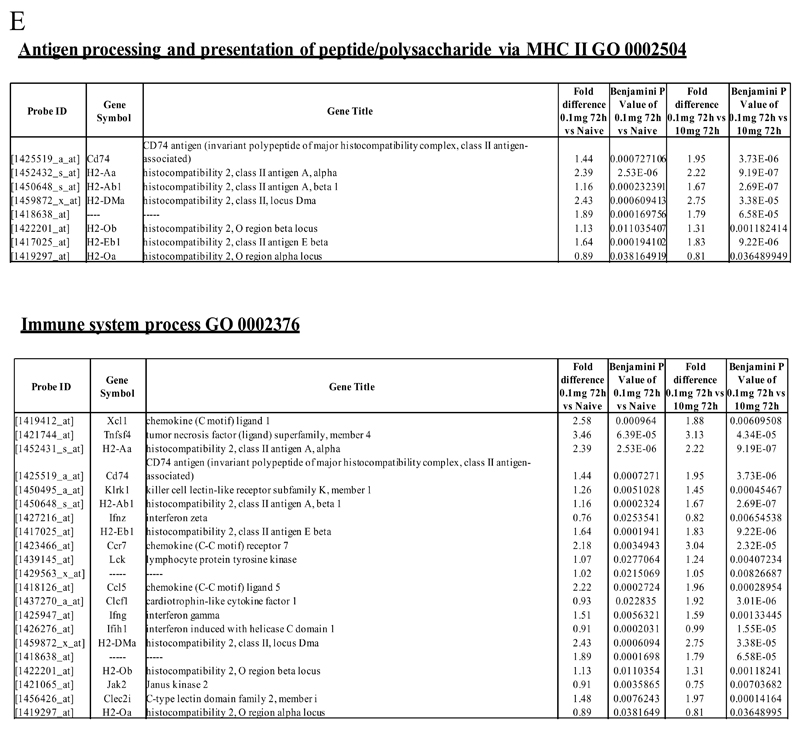
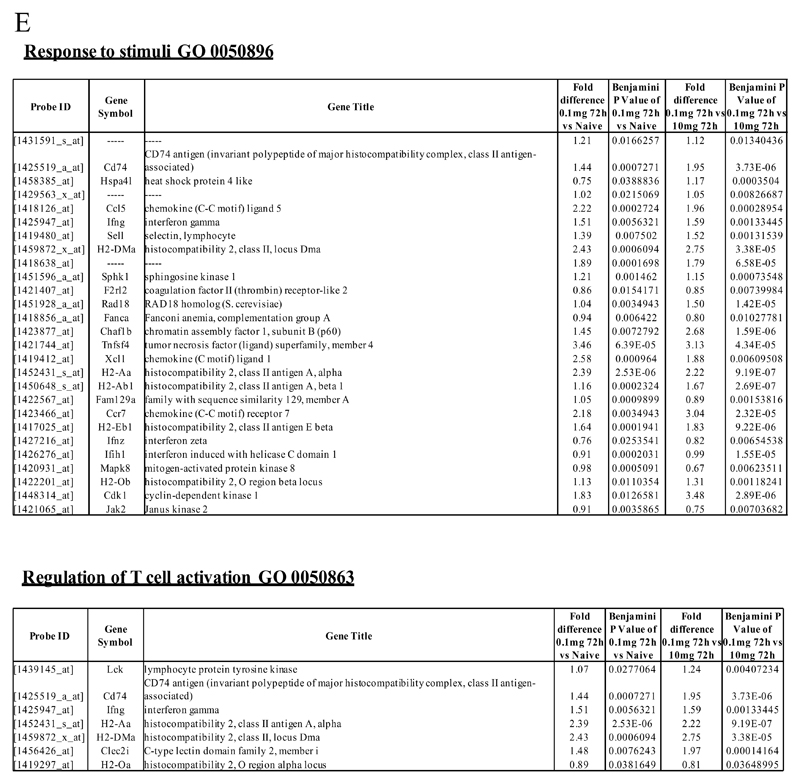
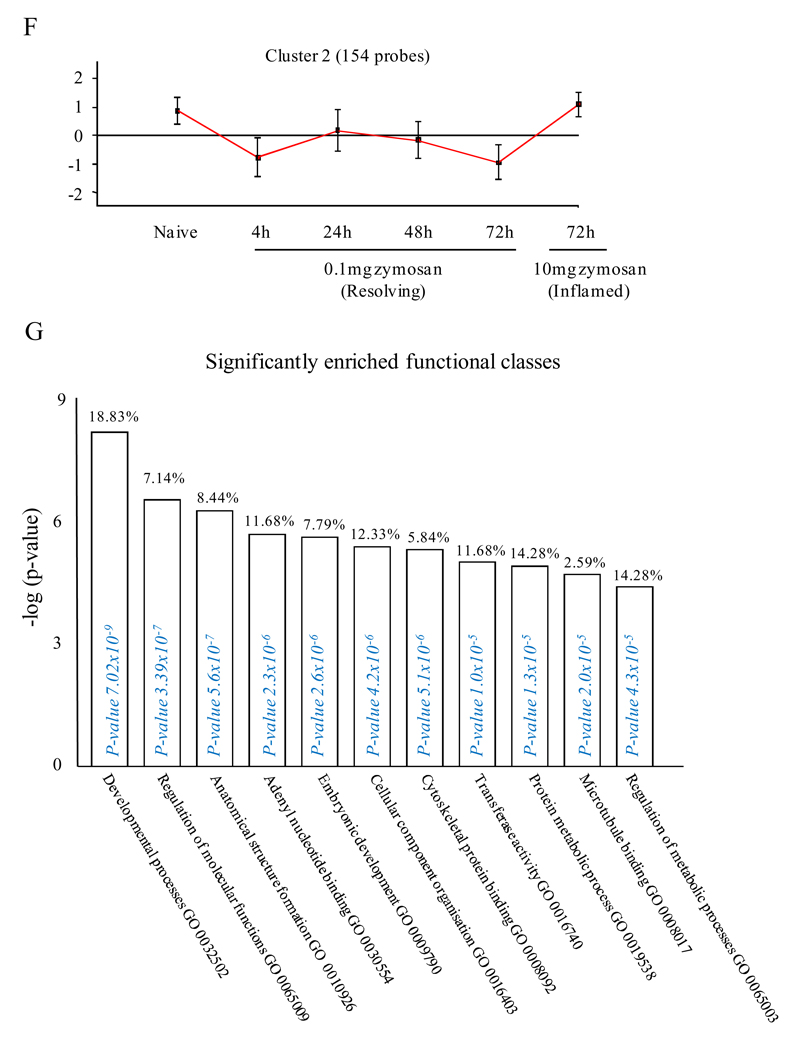
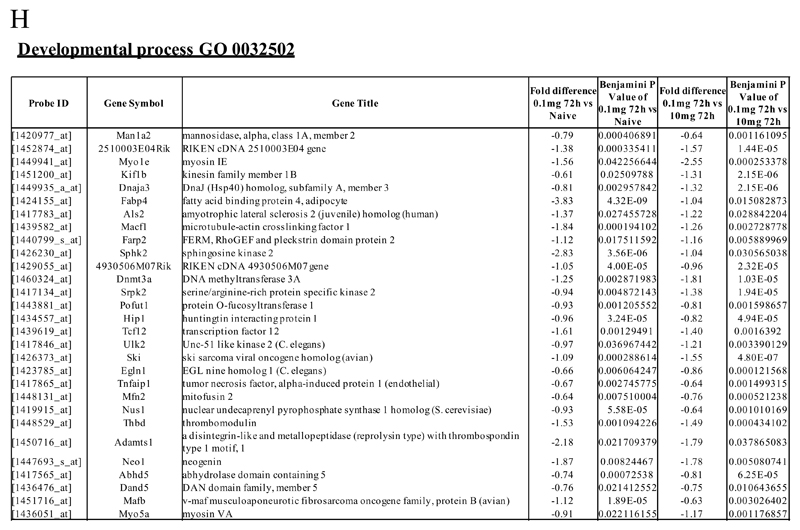
Figure 2. Genes enriched in rM cells compared to naive and pro-inflammatory macrophages.
An FDR of 0.05 and fold difference of 1.5 revealed (A) 342 genes up- and down-regulated in rM compared to pro-inflammatory (10mg zymosan) and naive macrophages, a sample of the most differentially expressed genes is presented in (B). The software package Expander detected (C) 191 probesets significantly up-regulated in rM cells with these gene sets enriched for (D) aspect of antigen uptake/presentation and immune function with a list of the genes involved in antigen processing and presentation of peptide/polysaccharide via MHC-II (GO 0002504), immune system process (GO 0002376), response to stimuli (GO 0050896) and regulation of T cell activation (GO 0050863) shown in (E). There was significant enrichment for (F) 154 down regulated probesets with their significantly enriched functional classes presented in (G). Genes involved in developmental processes (GO 0032502) are listed in (H).