Figure 4. Reproduction of data from the TCGA database with the cBioPortal tool.
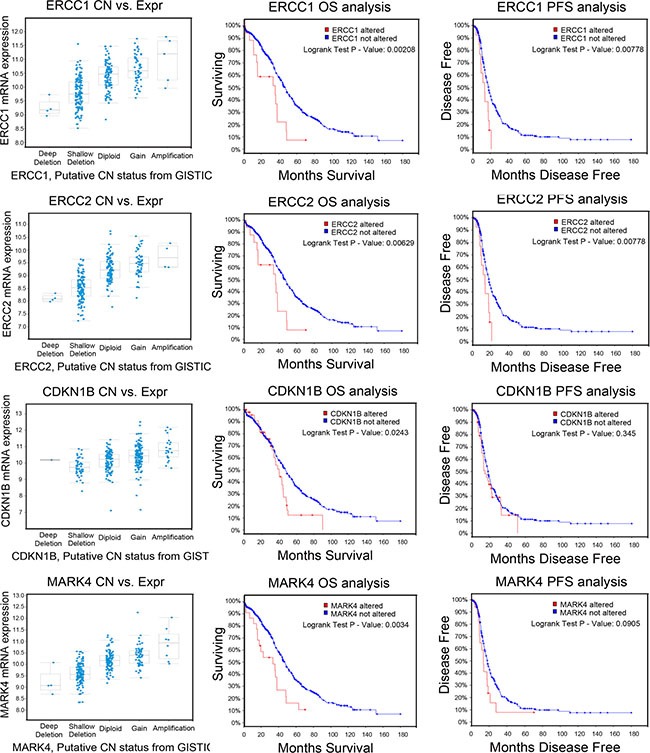
The amplifications of candidate genes, ERCC1, ERCC2, CDKN1B and MARK4 correlated with gene expression and were associated with patient outcomes. Kaplan-Meier analysis of OS and PFS was performed with the copy number of the candidate gene as a categorical variable, so that the effects of genes with unaltered and altered copy numbers could be compared. The results of a Cox proportional hazards test, with residual disease as a copredictor, are shown as P values. The correlations between the copy numbers and the mRNA levels of the candidate genes, determined through RNA Seq analysis of the TCGA datasets, are also shown.
