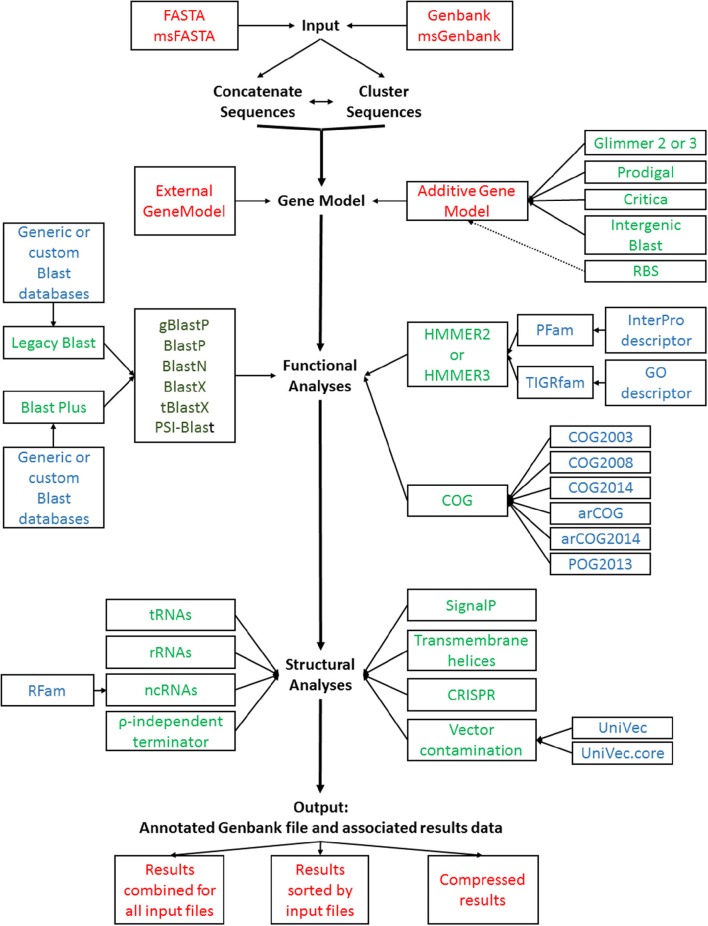
Figure 1.
GAMOLA2 annotation workflow. A schematic representation of the GAMOLA2 core annotation workflow. Input FASTA and Genbank sequences can be concatenated and/or clustered before submitted to gene model prediction, functional and structural analyses. The final output comprises an annotated Genbank file and associated data that can be viewed in Artemis or other suitable software. For convenience, the results may be compressed into a single archive file. Individual input or output files are shown in red (each analysis generates text output files well which are stored in their respective directories, not shown); programs are shown in green, available Blast flavors are shown in dark green; databases used are indicated in blue.