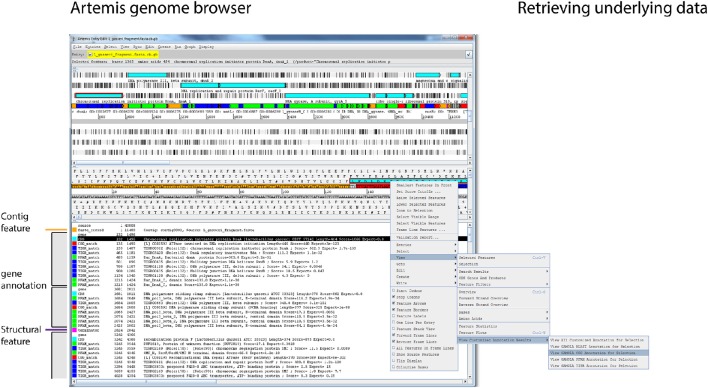
Figure 3.
Genome visualization in Artemis. Screenshot of the modified Artemis genome browser displaying a GAMOLA2 annotated sequence. The Artemis genome browser is a Java based application that is platform independent and can, once the Genbank file is loaded, traverse along the genome and display information for individual genes in real-time. Annotations for individual genes are presented in individual feature blocks that always begin with the “gene” and “CDS” features (gray boxes). Additional features are shown based on their respective genome location. Each feature has a defined color code, creating a consistent user experience. Changing gene annotations is achieved by modifying the “gene” qualifier in the “gene” and “CDS” features, whereby “gene” features display a short gene name and “CDS” features a verbose description (Supplemental Figure 9A). Names of functional domains are often cryptic and do not directly contribute to the deciphering of the biological role of a given gene. Each feature in a GAMOLA2 annotation therefore contains additional information to explain the respective biological role (where known) or provide additional qualitative details (Supplemental Figure 9B). Genes that lack a close characterized homolog or well-known domains often remain annotated as “conserved hypotheticals.” Investigating all functional and structural information above the selected thresholds often reveals common biological themes that lead to a putative annotation. The modified Artemis genome browser can retrieve the underlying full results for Blast, COG, PFam, and TIGRfam for each gene as long as the original file and folder structure is maintained (Supplemental Figure 9C).