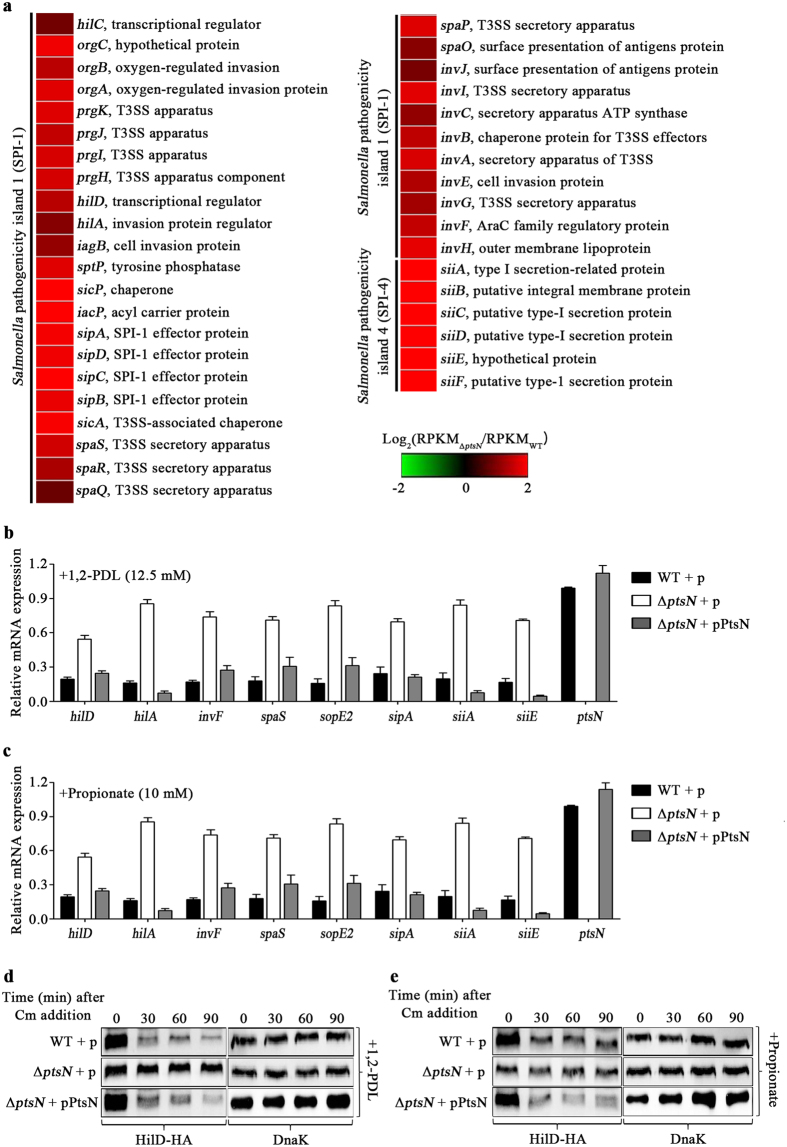
Figure 4. The effects of ptsN on the expression of genes involved in bacterial invasiveness.
(a) Heat-maps generated to describe the expression patterns of SPI-1 and SPI-4 genes in the ΔptsN mutant strain. Gene expression ratios between the ΔptsN mutant and wild-type SL1344 are shown using a colorimetric gradient: down-regulation in green and up-regulation in red. (b,c) The relative mRNA levels of the genes involved in SPI-1 and SPI-4, as measured by qRT-PCR. The tested genes are the SPI-1 regulator genes (hilD, hilA, and invF), SPI-1 genes (spaS, sopE2, and sipA), and SPI-4 genes (siiA and siiE). As alternative carbon sources, Ado-B12 (20 nM), 1,2-PDL (12.5 mM), and propionate (10 mM) were added to the cultures of (b and c), respectively. The mRNA level of each gene was normalized using that of gyrB in the wild-type strain containing pACYC184 and the ΔptsN mutant strains with either pACYC184 or pPtsN and its relative expression was calculated using the mRNA level of ptsN in the wild-type containing pACYC184, which was set at 1.0. All experiments were performed in triplicate, and the average values are depicted in the graphs. (d and e) The stability of the HilD-HA protein, compared under conditions identical to those used in (b) and (c). Three strains of wild-type with pACYC184 and ΔptsN mutant strains with either pACYC184 or pPtsN were cultivated in the presence of 1,2-PDL (d) or propionate (e). De novo protein synthesis was quenched by the addition of chloramphenicol (0.2 mg/ml) at 4 h post inoculation. Total proteins were harvested every 15 min, and equivalent amounts were subjected to SDS-PAGE analysis. The stability of HilD-HA was assessed using anti-HA antibody. The levels of DnaK as controls were comparable between lanes. Full-length blots were presented in Supplementary Figure S5.