Abstract
The treatment or cure of HIV infection by cell and gene therapy has been a goal for decades. Recent advances in both gene editing and chimeric antigen receptor (CAR) technology have created new therapeutic possibilities for a variety of diseases. Broadly neutralizing monoclonal antibodies (bNAbs) with specificity for the HIV envelope glycoprotein provide a promising means of targeting HIV-infected cells. Here we show that primary human T cells engineered to express anti-HIV CARs based on bNAbs (HIVCAR) show specific activation and killing of HIV-infected versus uninfected cells in the absence of HIV replication. We also show that homology-directed recombination of the HIVCAR gene expression cassette into the CCR5 locus enhances suppression of replicating virus compared with HIVCAR expression alone. This work demonstrates that HIV immunotherapy utilizing potent bNAb-based single-chain variable fragments fused to second-generation CAR signaling domains, delivered directly into the CCR5 locus of T cells by homology-directed gene editing, is feasible and effective. This strategy has the potential to target HIV-infected cells in HIV-infected individuals, which might help in the effort to cure HIV.
Keywords: chimeric antigen receptor, HIV, gene editing, cell therapy, CAR T cell
Rawlings, Wagner, and colleagues show that T cells expressing chimeric antigen receptors based on HIV-neutralizing antibodies can selectively clear HIV-infected cells in culture and demonstrate that the use of gene editing to protect therapeutic T cells from HIV infection can improve their efficacy.
Introduction
More than 30 million people are infected with HIV.1 Antiretroviral treatment (ART) dramatically decreases mortality2 but is expensive and inconvenient, and HIV-infected people still have an increased risk of malignancies,3 cardiovascular4 and neurologic5 disease, and shortened life expectancy.6 Therefore, a cure for HIV remains an important treatment goal.
Adoptive T cell therapy as a strategy to treat or cure HIV has been investigated for decades.7, 8, 9, 10, 11 Because residual HIV expression continues despite effective ART12, 13, 14, 15, 16 and is required for viral rebound, HIV-infected cells should theoretically be targetable by a T cell therapeutic agent. Several mechanisms are thought to be responsible for the apparent failure of autologous cytotoxic T lymphocytes (CTLs) to clear reactivated cells in HIV-infected individuals: HIV evolution prior to ART quickly selects for CTL escape mutations;17, 18, 19 HIV Nef mediates downregulation of major histocompatibility complex class I (MHC-I),20, 21 protecting HIV-infected cells from T cell receptor (TCR)-dependent CTL killing; and HIV-specific CTL responses may be limited by exhaustion22, 23 or peripheral immune tolerance.24, 25 Over the last decade, major advances have been made in engineering human T cells via introduction of chimeric antigen receptors (CARs) to enable specific lysis of pathogenic targets.26, 27, 28, 29 Because CAR T cell activity is MHC-independent, potent, and enforced by expression of an engineered gene cassette, anti-HIV CAR T cells might overcome the limitations of autologous CTLs. In fact, although the major clinical success has occurred in treatment of select lymphoid malignancies, one of the earliest CAR T cell therapies to reach clinical trials was designed for the treatment of HIV.8
The early attempts at CAR T cell therapy for HIV involved the adoptive transfer of T cells expressing a fusion of the human CD4 extracellular domain (a ligand of HIV) to the CD3ζ signaling domain.8 In a randomized trial, this first-generation CAR was safe and reduced the HIV reservoir as measured by a viral outgrowth assay, and CAR+ cells were detectable for 10 years despite the lack of clinical benefit.30, 31, 32 The efficacy of this approach was likely compromised because of limited CAR activity in the absence of an intracellular co-stimulatory signaling domain and the potential for HIV infection of T cells expressing the CD4 CAR.10, 33
A panel of novel high-affinity, broadly neutralizing monoclonal antibodies (bNAbs) recognizing the HIV envelope glycoprotein have been isolated and characterized over the last decade.34, 35 We predicted that single-chain variable fragments (scFvs) derived from these bNabs could be used to develop potent anti-HIV CARs (HIVCARs), and multiple scFvs could be selected to develop HIVCARs targeting different epitopes of the HIV envelope glycoprotein.
CCR5 is the primary co-receptor for HIV.36, 37 Individuals with an allelic variant that is not functional (CCR5Δ32) are protected from CCR5-tropic HIV infection.38 Hematopoietic stem cell transplant using a CCR5Δ32 donor led to the only known cure of HIV-1 infection,39, 40 and T cells treated with engineered nucleases that introduce mutations at the CCR5 locus are resistant to HIV,41, 42, 43, 44, 45 accelerating ongoing efforts to develop gene editing- and cell-based therapeutic agents for HIV.11, 46
Using new gene-editing techniques, it has recently become possible to achieve high rates of homology-directed recombination (HDR) of therapeutic cassettes into targeted loci, including CCR5 in primary T cells.47, 48, 49, 50 We have previously shown introduction of cDNA expression cassettes at the CCR5 locus in primary human T cells using an mRNA-delivered megaTAL nuclease and a homologous AAV donor template at rates of up to 60%.48 HDR has the potential advantage of simultaneous introduction of a CAR and disruption of CCR5 to protect engineered cells from HIV. Based on these combined rationales, the current study tested the concept that T cells utilizing CARs based on scFvs derived from high-affinity bNAbs and containing second-generation co-stimulatory domains, in parallel with genetic protection from HIV by disruption of CCR5, would be effective at targeting HIV-infected cells. Additionally, we evaluated the functional activity of T cells and achieved CCR5 disruption by delivery of the HIVCAR gene cassette into CCR5 via HDR.
Results
Construction of HIVCARs Derived from bNAbs Targeting Alternative Epitopes on the HIV Envelope Glycoprotein
HIV bNAbs are human antibodies isolated from HIV-infected donors that neutralize multiple HIV strains in vitro.34, 35 Hundreds of monoclonal bNAbs of varying breadth and potency have been identified and characterized in neutralization assays.51 We chose four high-breadth, high-potency bNAbs that bind different epitopes on the HIV envelope glycoprotein (Figure 1A): PGT-145 (variable regions 1 and 2 glycan loop), VRC07-523 (CD4-binding site), PGT-128 (mannose-rich region), and 10E8 (gp41 membrane-proximal external region).51, 52, 53, 54 To generate anti-HIVCARs, the heavy and light chains of each bNAb were synthesized as an scFv and cloned into a lentivirus (LV) second-generation CAR expression construct; blue fluorescent protein (BFP) was co-expressed downstream of a self-cleaving peptide (Figure 1B). An anti-CD19 scFv CAR (CD19CAR) was used as a control.
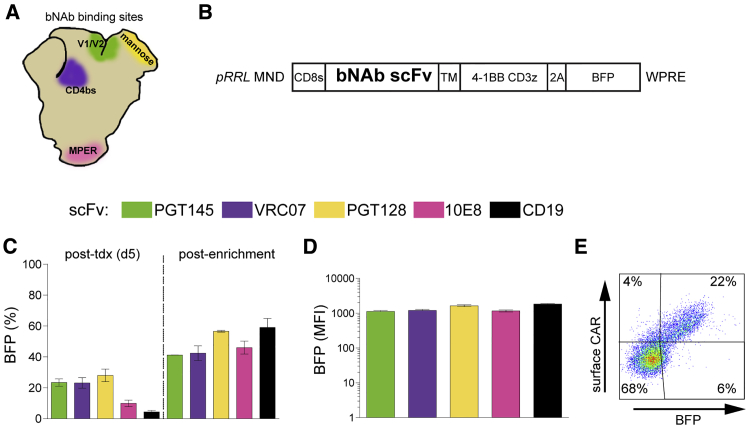
Figure 1.
HIVCARs Based on bNAb Are Expressed on the Surface of Primary Human T Cells
(A) Known binding site for each bNAb scFv used indicated by color on a diagram of the HIV envelope. V1/V2, variable loops 1 and 2; mannose, high-mannose patch; CD4bs, CD4 binding site; MPER, membrane proximal external region. (B) Schematic diagram of the CAR construct in the pRRL LV backbone containing the γ-retrovirus-derived promoter-enhancer MND.65 scFvs from various bNAbs (indicated by colored boxes below) were cloned upstream of the hinge region. CD8s, CD8-signaling domain; TM, CD8 trans-membrane domain; 4-1BB CD3z, intracellular signaling domains of second-generation CAR;64 2A, self-cleaving 2A peptide. (C) Percentage of BFP+ human primary CD3+ cells 5 days after LV transduction (tdx), and 8 days after enrichment by fluorescence-activated cell sorting (FACS). (D) MFI of BFP+ cells 8 days after enrichment. The bars in (C) and (D) show the mean ± SEM of n = 3 human cell donors. The same three donors were used for replicate transduction of each LV. (E) Representative flow plot showing surface CAR expression on primary human T cells transduced with pRRL MND VRC07-523-CAR T2A BFP.
Initial transduction of HIVCAR LVs at MOI ∼2 in primary human CD3+ cells produced 7%–20% positive cells (Figure 1C). Although much higher levels of T cell transduction were achievable with our LV constructs, a low MOI was utilized in our experiments to permit assessment of functional activity of each construct in cells with ∼1 viral integration/cell and, thus, limit variability that might be caused by variations in cell surface expression. The CD3+ cells used were obtained from three unique donors. T cells from each donor were transduced with all four HIVCAR LVs or the control CD19CAR LV in parallel to allow discrimination between donor T cell versus HIVCAR variations. T cells were sorted on BFP to enrich for transduced cells and match expression levels between HIVCAR T cell populations. Eight days after sort enrichment, expression was stable at 42%–58% BFP+ (Figure 1C). Differences in HIVCAR expression by BFP mean fluorescence intensity (MFI) were not significant between the constructs at this time point (Figure 1D). To confirm that the scFvs were expressed at the cell surface, cells transduced with the VRC07-523-HIVCAR and CD19CARs, which have kappa light chains, were stained with Protein L, demonstrating a linear correlation of Protein L staining with BFP expression, as would be expected, because of their cis linkage via a 2A sequence (Figure 1E).55
Specific Activation and Cytotoxic Activity of HIVCAR T Cells
CAR T cell function can be assayed by the detection of T cell activation markers on CAR+ cells after co-culture with cells that express the target antigen. BFP+ HIVCAR T cells expressed increased CD137 after 24 hr of co-culture with a stably infected HIVpos T cell line in the presence of ART (Figure 2A). In co-culture with HIVpos T cells, CD137 expression on BFP+ HIVCAR T cells was upregulated on a significantly higher percentage of cells than that of control BFP+ CD19CAR T cells for all scFvs tested, except for PGT128 HIVCAR T cells (Figure 2B). Parental HIVneg cells did not elicit CD137 responses. The difference in percentage of CD137-expressing BFP+ T cells after culturing with HIVpos versus HIVneg cell lines was significant for all HIVCAR T cells tested but not for the anti-CD19CAR control. The increase in the percentage of cells expressing CD137 after stimulation with HIVpos versus HIVneg targets cells was on average 7- to 8-fold with the VRC07-523, PGT145, and 10E8 CAR T cells and ∼3-fold with the PGT128 CAR, with some variability between the three T cell donors (Figure S1A). BFP+ HIVCAR T cells produced IL-2 and interferon γ (IFNγ), detectable by intracellular staining in co-culture with cell lines expressing a target HIV envelope (Figures S1B–S2D).
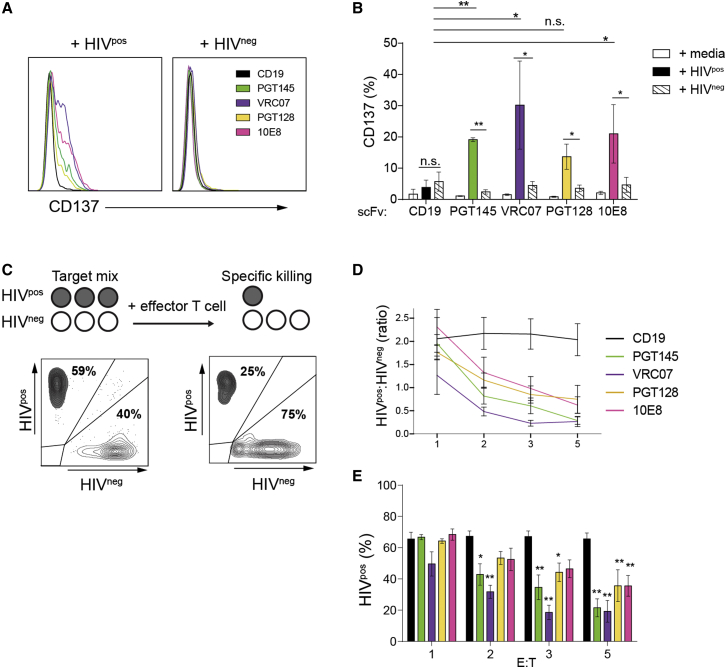
Figure 2.
Responses of HIVCAR T Cells to HIV-Infected Cells Are scFv-Specific
(A) Histograms showing CD137 expression on BFP+ CAR T cells 24 hr after mixing with either HIVpos or HIVneg target T cell lines. Shown are representative data from one of three independent donors. (B) Summary of CD137 expression in BFP+ CAR T cells generated from three donors incubated either with medium alone or HIVpos or HIVneg target T cell lines. (C) Schematic of the cytotoxicity assay and representative plots of two target cell populations before and after HIVCAR T cell treatment. (D) Ratio of HIVpos to HIVneg live target cells at 48 hr in wells plated with PGT145, VRC07-523, PGT128, 10E8, and CD19CAR T cells at increasing E:T ratios. (E) Percent of HIVpos target cells of total live target cells remaining 48 hr after plating with effector CAR T cells at increasing E:T ratios. The significance shown is versus anti-CD19CAR at each E:T ratio. For all charts, bars show the mean ± SEM of three unique experiments. p values were obtained using the Tukey method for ANOVA for multiple comparisons. *p < 0.05; **p < 0.001; n.s., not significant.
HIVCAR T cells were next tested for their capacity to specifically kill HIV-infected cells in a mixed background of uninfected cells. Fluorescently labeled HIVpos cells were mixed with HIVneg cells and then cultured with various CAR T cells in the presence of ART (Figure 2C). Increasing doses of HIVCAR T cells led to greater reductions in the ratio of HIVpos to HIVneg cell targets 48 hr post-mixing (Figure 2D). At a 5:1 effector-to-target (E:T) ratio, the percentage of HIVpos target cells decreased by 43% with PGT128 or 10E8 HIVCAR T cells and 70%–72% with VRC07-523 or PGT145-HIVCAR T cells compared with CD19CAR T cells. Although no HIVCAR was significantly different from another at any E:T ratio, only the VRC07-523 and PGT145-HIVCARs showed a significant relative decrease in HIVpos cells at a 2:1 E:T ratio, and all except 10E8 had significant decreases at a 3:1 ratio and above (Figure 2E). In a live virus assay, all HIVCARs except 10E8 were able to control viral replication of the HIV-1JR-CSF strain, as measured by HIV capsid ELISA (Figure S2).
Of the scFv CARs tested, the PGT145- and VRC07-523-CARs appeared to show the most consistent potency in our initial assays. Interestingly, the VRC07-523-CAR targets a similar region of the HIV envelope as the early anti-HIV CD4-based CAR.53 Given the tendency for HIV to mutate in response to pressures, it is likely to be advantageous to develop anti-HIV CARs targeting multiple epitopes on the HIV envelope. The PGT145-CAR, targeting the V1/V2 glycan region, was therefore selected for testing in conjunction with CCR5 disruption.
Homology-Directed Repair for Targeted Integration of PGT145-HIVCAR at the CCR5 Locus
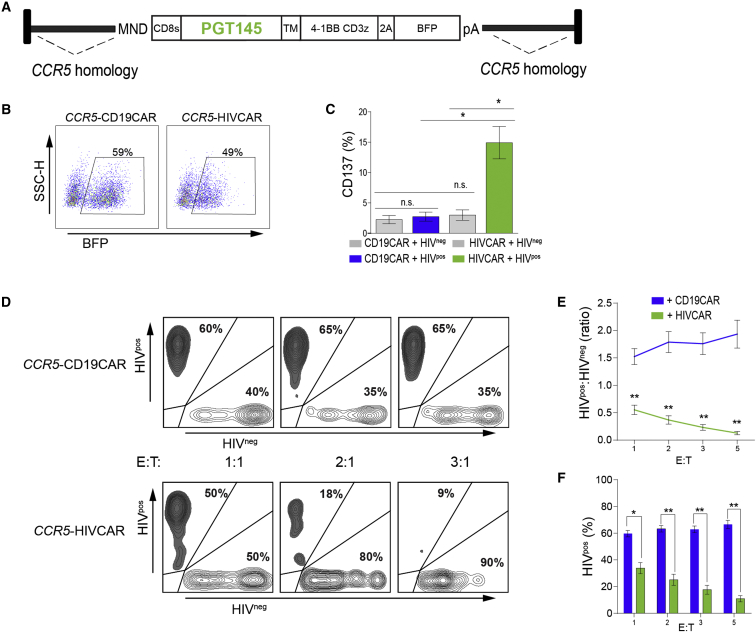
CD4+ T cells have an important role in maintaining the survival of cytotoxic lymphocytes in vivo, and recent studies with CAR T cells targeting lymphoma suggest that a defined mix of both CD4+ and CD8+ T cells may have higher efficacy and persistence in patients.31, 56, 57 However, CD4+ CAR T cells are expected to be vulnerable to HIV-1 infection, and there is evidence that CD8+ CAR T cells expressing the CD4-based CAR can be infected.10, 33 Because CCR5 is the predominant co-receptor used for HIV entry, we tested the effect of CCR5 disruption on HIVCAR T cell function. We have previously published the targeted integration of a CAR construct at the CCR5 locus with rates of 10%–15% in primary human T cells.48 Using the CCR5 megaTAL and an AAV donor template similar to that described previously (Figure 3A), we introduced the PGT145-HIVCAR at the CCR5 locus or the CD19CAR as a control. After sort enrichment for BFP+ cells, the resultant CCR5-HIVCAR (PGT145) T cells were specifically activated, as measured by cell surface CD137 expression at 24 hr, when stimulated with the HIV-infected (HIVpos) cell line (Figures 3B and 3C). CCR5-HIVCAR T cells also specifically killed HIV-infected cells in the presence of ART in a dose-dependent fashion, as measured by reduction in the ratio of HIVpos cells (GFP+) to HIVneg cells (mCherry+) and reduction in the percentage of HIVpos live target cells (Figures 3D–3F).
Figure 3.
Effector T Cells Generated by Targeted Integration of an HIVCAR Cassette into CCR5 Retain HIV-Specific Activation and Lysis Capacity
(A) Map of the AAV6 vector for HDR targeting of the PGT145-CAR expression cassette to human CCR5. CCR5 homology arms are approximately 0.6 kb. (B) Flow plots showing expression of cis-linked BFP in sort-enriched CCR5-CAR T cells used in functional assays. (C) Percentage of CCR5-HIVCAR T cells that express CD137 24 hr after stimulation with HIVpos (colored) or HIVneg (gray) T cell lines compared with activity of CCR5-CD19 CAR T cells in an identical setting. (D) Representative flow plots of the target cell mix 48 hr after plating at increasing E:T ratios with CCR5-CD19 or CCR5-HIVCAR T cells. Percentages shown are percent GFP+ (HIVpos target cell line) or mCherry+ (HIVneg target cell line) from a gate that excludes BFP+ (effector) and double-negative cells. (E) Ratio of HIVpos (GFP) to HIVneg (mCherry) live target cells at increasing E:T ratios, performed in triplicate. (F) Percentage of total target cells that are HIVpos as measured by GFP expression. The significance shown is a t test comparison of CD19CAR versus HIVCAR T cells at each E:T ratio. Shown are mean ± SEM for n = 5 (C) of three (E and F) unique experiments using CAR T cells generated from three independent human donors. p values calculated using unpaired two-tailed t test are indicated as follows: *p < 0.05, **p < 0.001.
CCR5-Edited HIVCAR T Cells More Effectively Suppress Viral Replication
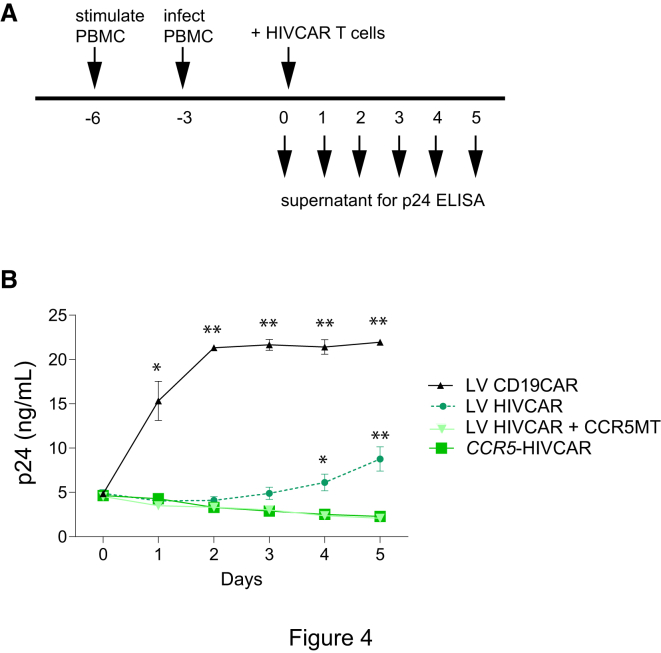
The efficacy of adoptive HIV CAR T cell therapy is expected to be challenging in the context of active viral replication. To test whether CCR5 disruption provides a meaningful benefit in this context, HIVCAR and control CD19CAR T cells with or without CCR5 modification were added to cultures containing peripheral blood mononuclear cells (PBMCs) with actively replicating HIV. Control of HIV replication was monitored daily by ELISA for the HIV capsid protein p24 (Figure 4A). HIVCAR T cells produced by three methods were directly compared: LV delivery of the PGT145-HIVCAR, LV delivery of the PGT145-HIVCAR plus megaTAL disruption of CCR5 via non-homologous end joining (NHEJ), and HDR using the CCR5 megaTAL plus AAV donor template with CCR5 homology arms (CCR5-PGT145HIVCAR). Through days 1–3 of co-culture with infected PBMCs, all three HIVCAR T cell products controlled viral replication significantly better than CD19CAR T cells, and there were no significant differences between the three types of HIVCAR T cells. However, on days 4–5, there was a significant increase in viral particles when HIVCAR T cells without CCR5 disruption were used (Figure 4B). These findings support the idea that, in the presence of active virus replication, CCR5 disruption is likely to be beneficial and/or critical for effective HIVCAR T cell therapy.
Figure 4.
CCR5 Disruption of HIVCAR T Cells Improves Viral Control In Vitro
(A) Live virus challenge of CAR T cells during a 5-day co-culture with HIV-infected allogenic PBMCs. (B) p24 concentration in supernatant plotted over time. p values were obtained using the Tukey method for ANOVA for multiple comparisons. The significance shown is a comparison of each condition versus CCR5-HIVCAR. *p < 0.05, **p < 0.001. Error bars show ± SEM, n = 4 (each n is the average of duplicate samples) using T cells from two donors.
Discussion
Our studies lay an important foundation for further development of anti-HIV CAR adoptive cellular therapies. We demonstrate robust anti-HIV activity of four distinct second-generation CARs derived from bNAbs. Each CAR tested used an scFv targeting a distinct epitope on the gp120 glycoprotein, demonstrating that bNAbs of multiple classes are compatible with CAR technology. We also demonstrate that HIVCAR T cells exhibit activity toward HIV-infected cell lines in the presence of ART. Each of the bNAb-based HIVCAR expression constructs tested induced specific T cell activation and killing of HIV-infected cells compared with control CD19CAR T cells, and no HIVCAR T cell activation occurred in culture with the uninfected cell line. These mixed target cell cultures grown in the presence of ART were designed to mimic the anticipated scenario encountered during clinical usage of HIVCAR T cells; clearance of HIV-infected cells in these assays demonstrates the potential of HIV CARs to target and eliminate HIV-infected cells in individuals on ART.
Among the candidate HIVCARs evaluated, the PGT145 and VRC07-523-HIVCARs appeared to be the most effective in our studies, exhibiting efficient CD137 upregulation and cytokine production, killing at low effector to target ratios, and sustained efficacy in the setting of CCR5 disruption (for PGT145). PGT128 CAR T cells, in contrast, exhibited lower rates of CD137 expression after co-culture with HIVpos targets and appeared to clear HIVpos cells less well than other HIVCAR T cells but produced cytokines after stimulation with cells expressing an alternative HIV envelope. Ali et al.58 recently compared the potency of bNAb-based HIV CARs against HIV-1NL4-3-infected T2 cells and found the CAR based on the PGT128 bNAb to be the least effective, whereas that based on the 10E8 bNAb was the most potent. Here we observed that 10E8 HIVCAR T cells exhibited an activation profile that varied between donors and killed less potently than PGT145- and VRC07-523-HIVCAR T cells in our target cell assays. The 10E8 antibody binds at the membrane-proximal external region (MPER), which may be less accessible than other regions of the HIV envelope. In a live-virus assay, the 10E8 HIVCAR T cells were also the only HIVCAR cells tested that failed to control HIVJR-CSF replication, as measured by supernatant p24. Although 10E8 has been shown to neutralize JR-CSF efficiently,52 our finding is perhaps not surprising given the observation that anti-MPER antibodies bind poorly to the surface of infected cells.59 The potency of different HIVCARs against different viral variants supports the pursuit of HIVCAR combinations. Our experiments were designed to test the feasibility of using bNAbs to generate multiple CARs for a potentially combinatorial approach for HIV therapy in the clinic rather than to specifically compare the effectiveness of targeting a particular region of the HIV envelope. Additionally, even with the most studied CD19CAR, it is not yet clear which in vitro assays best predict clinical success.26 Therefore, we hesitate to make definitive claims regarding the relative performance of candidate HIVCARs derived from the alternative bNAb scFvs. Given the propensity of HIV to mutate to evade specific pressures, it seems prudent to develop multiple HIVCARs that target different epitopes and could be used in combination to treat HIV.
An important additional advance in this study is the use of gene editing to generate HIV-resistant HIVCAR T cells. CD8+ CAR T cells expressing a CD4-based CAR have previously been shown to be susceptible to HIV infection,33 and it is likely that CD4+ T cells expressing this construct might be even more susceptible to infection. It is conceivable that this was a key limitation of the HIVCAR T cells studied in previous randomized trials.30, 31 We show that it is possible to protect HIVCAR T cells by independent disruption of CCR5 by NHEJ or HDR. Both methods produce functional CAR T cells that kill HIV-infected cells in the presence of ART, and both types of HIV-resistant CAR T cells outperformed HIVCAR T cells without CCR5 disruption in live viral assays. Either gene-editing method could be used to generate further preclinical data. Ex vivo gene editing of CCR5 by NHEJ has been shown to be safe and effective in a phase I clinical trial,44 whereas HDR is a newer technology that has not yet been tested clinically. However, HDR has a potential advantage in that it limits the number of possible integration events and may reduce the risk of insertional mutagenesis. In addition, because of the nature of the simultaneous CAR insertion/CCR5 disruption, enriching for CAR+ cells generated through HDR also enriches for cells with CCR5 disruption. Finally, compared with LV delivery of the CAR, HDR has the added advantage that it can be done in the presence of ART, which may be an important safety feature during ex vivo manipulation of cells from HIV-infected individuals. Interestingly, we observed a trend for increased killing efficiency of HIVpos target cells in experiments using CCR5-PGT145-HIVCAR compared with LV-delivered PGT145-HIVCAR, particularly at lower effector-to-target cell ratios (Figures 2E and 3F). It will be important to directly compare these cell products in additional in vivo and in vitro assays to determine whether this observation has functional relevance.
Although our in vitro data on CCR5-disrupted HIVCAR T cells highlight the promise of a CAR T cell approach to eradication of HIV, several potential in vivo challenges exist. One concern is the potential to induce a cytokine release syndrome (CRS). CRS and associated neurological toxicities are seen in 10%–20% of patients treated with CD19CAR T cells, and studies of these patients suggest that cell dose, conditioning regimen, CAR T cell composition, and, particularly, tumor burden influence the risk of CRS.60, 61 The number of HIV-infected target cells in patients on ART is dramatically less than target cell numbers in individuals with leukemia. Consequently, the risk for HIVCAR-induced CRS would be predicted to be lower.
A significant challenge regarding the efficacy of HIVCAR T cell therapy is the requirement for persistent activity. Mathematical modeling suggests that a 5-year HIV-free period may be necessary to establish an effective cure.62 The CAR T cell dose, composition, phenotype, and immune-conditioning regimens will all likely influence CAR T cell persistence. Another potential concern is that HIVCAR constructs might be immunogenic. Although CAR constructs are assembled from human protein domains, the junctions between these domains may be seen as foreign, and, therefore, may require additional optimization. Persistent activity also requires preventing HIV Env escape. In the absence of ART, HIV has an extremely high mutation rate, allowing rapid selection for resistance mutations, which would likely include resistance to a single scFv-based CAR. We envision addressing this concern by adoptive transfer of anti-HIV CAR T cells to individuals who are initially on ART and by combining CAR T cells targeting distinct epitopes on HIV Env, potentially in concert with agents that promote viral activation in latently infected cells.63
In summary, we have demonstrated the feasibility and utility of a novel strategy for HIVCAR therapy that not only targets HIV-infected cells in the presence of ART but also protects the effector cells from HIV infection. This approach leverages recent advances in the ability to identify potent bNAbs, CAR technology, and the field of gene therapy, including the ability to perform high-efficiency gene editing of primary cells, and warrants further preclinical testing in animal models.
Materials and Methods
CAR Design and Vector Cloning
A plasmid with a T7 promoter driving the CCR5 megaTAL coding sequence and pRRL-MND-CD19CAR-BFP have been described previously.48 HIV scFvs were exchanged with that of the CD19 scFv in this plasmid as follows. scFvs from the previously reported human anti-HIV bNAbs PGT145, PGT128, 10E8, and VRC07-52351, 52, 53 were gene-synthesized by GenScript (Figure S3). Synthesized scFvs included additional sequences for cloning into BamHI and NheI sites of pRRL-MND-CD19CAR-BFP while maintaining the reading frame with upstream human CD8 signal peptide and downstream CAR domains (including CD8α hinge and transmembrane domains and 4-1BB and CD3ζ intracellular signaling domains64). The AAV6-CCR5-HIVCAR plasmid is as described previously,48 except that RQR8 was replaced by BFP. Constructs were verified by restriction digestion and sequencing. For LVs used to transduce target cell lines, either an mCherry or GFP fluorophore was cloned between an MND promoter65 and a woodchuck hepatitis virus post-transcriptional response element (WPRE) in the pRRL backbone. Viral LV (vesicular stomatitis virus G protein [VSV-G] pseudotyped) and AAV6 were produced, and titers were determined as described previously.48
Primary and Transformed T Cell Cultures
Primary CD3+ T cells were isolated from the peripheral blood of healthy donors (with approval of the Seattle Children’s Research Institute’s Institutional Review Board) by negative selection using the RosetteSep human T Cell kit (STEMCELL Technologies). ACH-266, 67 and A3.0168, 69 cell lines were obtained from Dr. Thomas Folks through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH. Human PBMCs, the CD34-depleted flowthrough from granulocyte-colony stimulating factor (G-CSF)-mobilized apheresis, were obtained from the Fred Hutchinson Cancer Research Center Hematopoietic Cell Processing and Repository core. All T cells and PBMCs were cultured in RPMI 1640 medium with 20% fetal calf serum (Omega Scientific), 1× GlutaMAX, 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), and 55μM 2-mercaptoethanol (Gibco) and incubated in a humidified environment at 37°C (except where noted) with 5% CO2. Cells were maintained at a density of ∼1 × 106 cells/mL by expansion into larger culture volumes every 2–3 days. Primary T cell cultures were supplemented with the following recombinant human cytokines from PeproTech: interleukin-2 (IL-2, 50 ng/mL), IL-7 (5 ng/mL), and IL-15 (5 ng/mL). Primary PBMC cultures were supplemented with IL-2 (50 ng/mL, PBMC medium).
Generation of CAR-Modified T Cells by Lentiviral Gene Transfer or Gene Editing
For gene transfer using LVs, 1× 106 primary human T cells were plated at 2 × 106/mL in the presence of 4 μg/mL Polybrene (Sigma-Aldrich) and 10 μL (∼2 MOI) of LV. Cells were incubated overnight, and then an additional 1 mL medium was added. For gene editing, CAR expression cassettes were targeted to the CCR5 locus using a CCR5 megaTAL nuclease followed by AAV6-CCR5.CAR.BFP as described previously.48 Briefly, T cells (1 × 106/mL) were stimulated at a 1:1 ratio with CD3/CD28 beads (Dynal Beads, Thermo Fisher Scientific) for 48 hr, washed, and incubated at 5 × 105/mL without beads for 16 hr. 2.5–3 × 105 cells were then transfected with 1 μg CCR5 megaTAL mRNA (generated as described previously)48 using the Neon Transfection System and a 10 μL tip at the following settings: 1,400 V, 10 ms, 3 pulses. Electroporated cells were immediately transferred to 200 μL pre-warmed T cell medium in a 96-well plate and incubated at 30°C for 22–24 hr, followed by standard culture conditions at 37°C. During the 30°C incubation, 2–4 hr after electroporation, 40 μL (∼1 × 105 MOI) of AAV.CCR5.CD19-CAR.2A.BFP or AAV.CCR5.PGT145-CAR.2A.BFP was added to the cell medium.
Expression of viral transgenes (BFP+) and surface expression of the CD19 and VRC07-523-CARs, detected by staining cells with biotinylated Protein L (GenScript) and phycoerythrin (PE)-streptavidin (BD Biosciences), was confirmed by flow cytometry on an LSR II (BD Biosciences) 5–10 days after LV transduction or gene editing. One week after LV transduction or 2 weeks after gene editing, cells were sorted on a FACSAria I (BD Biosciences) to enrich for BFP+ cells. Stable BFP marking was assessed 6–8 days after sorting. 20 days after either LV transduction or gene editing, sorted cells were frozen in medium with 10% DMSO. Prior to use in downstream assays, pre-enriched frozen CAR T cells were thawed and expanded for 1 week in the presence of OKT-3 antibody, irradiated TM-LCL, and PBMCs as described previously.70
Activation and Cytotoxicity Assays
CAR T cell functions were tested using the ACH-2 and A3.01 cell lines. ACH-2 cells are stably infected with HIV-1 and are designated below as HIVpos cells. A3.01, the parental cell line of ACH-2, is a T cell line derived from an acute lymphocytic leukemia (ALL) patient and is designated below as HIVneg cells. Using the method described above for LV transduction of primary T cells, HIVpos cells were transduced with LV-MND.GFP and HIVneg cells with LV-MND.mCherry at MOI ∼20. 5 days after LV addition, transduction was measured by flow cytometry as >99% for the relevant fluorophore. Prior to each assay, both HIVpos and HIVneg cell lines were stimulated with 1 μM phorbol-12-myristate 13-acetate (PMA) in the presence of a triple ART cocktail (20 μM each tenofovir, zidovudine, and nevirapine, each obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH). 24 hr later, cells were washed three times in PBS and resuspended at 1 × 106/mL in T cell medium with cytokines and the triple ART cocktail.
For CAR T cell activation assays, 5 × 104 target cells (either HIVpos or HIVneg) were plated in each well of a 96-well plate with 1 × 105 CAR-modified T cells. Cells were incubated for 24 hr and then stained for viability (Live/Dead Fixable Near-IR Kit, Thermo Fisher Scientific) and CD137 (PE, BD Biosciences), incubated for 15 min in 4% paraformaldehyde, and analyzed by flow cytometry. Activation of CAR T cells was reported after gating on BFP+ live lymphocytes.
For target cell killing assays, we plated a 1:1 mix of HIVpos and HIVneg cells (5 × 104 each, 1 × 106/mL) in T cell medium containing cytokines and ART into individual wells of a 96-well plate. CAR-modified T cells were then added (5 × 104, 1 × 105, 1.5 × 105, or 2.5 × 105 for a 1:1, 2:1, 3:1, or 5:1 effector-to HIVpos-target cell ratio, respectively). After culturing the mixture for 48 hr, cells were stained for viability and fixed with paraformaldehyde, followed by flow cytometry as described above.
Live-Virus Assays
HIV-1JR-CSF71, 72 was obtained from Dr. Irvin Chen through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. The ability of CAR T cells to control HIV-1 infection in a cell culture model was assayed using methods adapted from previously published work.73, 74 PBMCs were cultured for 72 hr in PBMC medium with 2 μg/mL phytohemagglutinin (PHA, Sigma-Aldrich). The medium was then removed, and cells were re-plated at 5 × 106/mL in 6 mL PBMC medium, and HIV-1JR-CSF was added to the PBMC culture at an MOI of 1 × 10−3. After 2-hr incubation, the medium was removed, and the infected PBMCs were then incubated in fresh PBMC medium at 1 × 106/mL for an additional 72 hr. After 72 hr, infected PBMCs were diluted to 5 × 105 cells/mL in T cell medium with cytokines. ∼2.5 × 104 infected PBMCs (50 μL) were co-cultured with ∼5 × 104 CAR T cells (100 μL) per well in a 96-well plate. 30 μL supernatant was removed from wells and frozen 1 hr after plating and then every 24 hr for the next 3 days; this volume was replaced with an equivalent volume of fresh T cell medium with cytokines. Viral antigen p24 concentrations were then quantified in supernatant fractions by p24 ELISA (Zeptometrix). Each condition was run in duplicate reactions, and each reaction was assayed in duplicate ELISAs.
Data Analysis
Flow cytometry data were analyzed using FlowJo software version 9.2 or 10.7 (Tree Star). Statistical analyses were performed using GraphPad Prism 6 (GraphPad). Tests of statistical significance were performed using unpaired two-tailed Student’s t test or ANOVA.
Author Contributions
M.H. designed and performed research, contributed reagents, analyzed data, and wrote the paper. T.M., G.S.R.I., J.S., and A.B. developed assays, performed research, and analyzed data. K.S. contributed reagents and wrote the paper. A.M.S., D.J.R., and T.A.W. designed research and wrote the paper.
Conflicts of Interest
A.M.S. is a consultant and shareholder in bluebird bio and receives compensation from bluebird bio. The remaining authors declare no competing financial interests. Seattle Children’s Hospital has filed patent applications related to this work.
Acknowledgments
We thank bluebird bio for providing the CCR5 megaTAL. This work was supported by the Seattle Children’s Research Institute Center for Immunity and Immunotherapies and Program for Cell and Gene Therapy, the Children’s Guild Association Endowed Chair in Pediatric Immunology (to D.J.R.), and the NIAID of the NIH under awards P30 AI027757 (to T.A.W.) and AI118500 (to T.A.W.).
Footnotes
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.12.023.
Contributor Information
David J. Rawlings, Email: drawling@uw.edu.
Thor A. Wagner, Email: thor.wagner@seattlechildrens.org.
Supplemental Information
References
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS) (2016). Global AIDS Response Progress Reporting. https://aidsreportingtool.unaids.org/static/docs/GARPR_Guidelines_2016_EN.pdf.
- 2.Palella F.J., Jr., Baker R.K., Moorman A.C., Chmiel J.S., Wood K.C., Brooks J.T., Holmberg S.D., HIV Outpatient Study Investigators Mortality in the highly active antiretroviral therapy era: changing causes of death and disease in the HIV outpatient study. J. Acquir. Immune Defic. Syndr. 2006;43:27–34. doi: 10.1097/01.qai.0000233310.90484.16. [DOI] [PubMed] [Google Scholar]
- 3.Deeken J.F., Tjen-A-Looi A., Rudek M.A., Okuliar C., Young M., Little R.F., Dezube B.J. The rising challenge of non-AIDS-defining cancers in HIV-infected patients. Clin. Infect. Dis. 2012;55:1228–1235. doi: 10.1093/cid/cis613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Triant V.A., Josephson F., Rochester C.G., Althoff K.N., Marcus K., Munk R., Cooper C., D’Agostino R.B., Costagliola D., Sabin C.A. Adverse outcome analyses of observational data: assessing cardiovascular risk in HIV disease. Clin. Infect. Dis. 2012;54:408–413. doi: 10.1093/cid/cir829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mothobi N.Z., Brew B.J. Neurocognitive dysfunction in the highly active antiretroviral therapy era. Curr. Opin. Infect. Dis. 2012;25:4–9. doi: 10.1097/QCO.0b013e32834ef586. [DOI] [PubMed] [Google Scholar]
- 6.Harrison K.M., Song R., Zhang X. Life expectancy after HIV diagnosis based on national HIV surveillance data from 25 states, United States. J. Acquir. Immune Defic. Syndr. 2010;53:124–130. doi: 10.1097/QAI.0b013e3181b563e7. [DOI] [PubMed] [Google Scholar]
- 7.Romeo C., Seed B. Cellular immunity to HIV activated by CD4 fused to T cell or Fc receptor polypeptides. Cell. 1991;64:1037–1046. doi: 10.1016/0092-8674(91)90327-u. [DOI] [PubMed] [Google Scholar]
- 8.Yang O.O., Tran A.C., Kalams S.A., Johnson R.P., Roberts M.R., Walker B.D. Lysis of HIV-1-infected cells and inhibition of viral replication by universal receptor T cells. Proc. Natl. Acad. Sci. USA. 1997;94:11478–11483. doi: 10.1073/pnas.94.21.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masiero S., Del Vecchio C., Gavioli R., Mattiuzzo G., Cusi M.G., Micheli L., Gennari F., Siccardi A., Marasco W.A., Palù G., Parolin C. T-cell engineering by a chimeric T-cell receptor with antibody-type specificity for the HIV-1 gp120. Gene Ther. 2005;12:299–310. doi: 10.1038/sj.gt.3302413. [DOI] [PubMed] [Google Scholar]
- 10.Zhen A., Kamata M., Rezek V., Rick J., Levin B., Kasparian S., Chen I.S., Yang O.O., Zack J.A., Kitchen S.G. HIV-specific Immunity Derived From Chimeric Antigen Receptor-engineered Stem Cells. Mol. Ther. 2015;23:1358–1367. doi: 10.1038/mt.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C.X., Cannon P.M. The clinical applications of genome editing in HIV. Blood. 2016;127:2546–2552. doi: 10.1182/blood-2016-01-678144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furtado M.R., Callaway D.S., Phair J.P., Kunstman K.J., Stanton J.L., Macken C.A., Perelson A.S., Wolinsky S.M. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 13.Santosuosso M., Righi E., Lindstrom V., Leblanc P.R., Poznansky M.C. HIV-1 envelope protein gp120 is present at high concentrations in secondary lymphoid organs of individuals with chronic HIV-1 infection. J. Infect. Dis. 2009;200:1050–1053. doi: 10.1086/605695. [DOI] [PubMed] [Google Scholar]
- 14.Palmer S., Maldarelli F., Wiegand A., Bernstein B., Hanna G.J., Brun S.C., Kempf D.J., Mellors J.W., Coffin J.M., King M.S. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasternak A.O., Jurriaans S., Bakker M., Prins J.M., Berkhout B., Lukashov V.V. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS ONE. 2009;4:e8490. doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatano H., Delwart E.L., Norris P.J., Lee T.H., Neilands T.B., Kelley C.F., Hunt P.W., Hoh R., Linnen J.M., Martin J.N. Evidence of persistent low-level viremia in long-term HAART-suppressed, HIV-infected individuals. AIDS. 2010;24:2535–2539. doi: 10.1097/QAD.0b013e32833dba03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., McNevin J.P., Holte S., McElrath M.J., Mullins J.I. Dynamics of viral evolution and CTL responses in HIV-1 infection. PLoS ONE. 2011;6:e15639. doi: 10.1371/journal.pone.0015639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore C.B., John M., James I.R., Christiansen F.T., Witt C.S., Mallal S.A. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 19.Deng K., Pertea M., Rongvaux A., Wang L., Durand C.M., Ghiaur G., Lai J., McHugh H.L., Hao H., Zhang H. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz O., Maréchal V., Le Gall S., Lemonnier F., Heard J.M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 21.Collins K.L., Chen B.K., Kalams S.A., Walker B.D., Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 22.Mueller Y.M., De Rosa S.C., Hutton J.A., Witek J., Roederer M., Altman J.D., Katsikis P.D. Increased CD95/Fas-induced apoptosis of HIV-specific CD8(+) T cells. Immunity. 2001;15:871–882. doi: 10.1016/s1074-7613(01)00246-1. [DOI] [PubMed] [Google Scholar]
- 23.Petrovas C., Chaon B., Ambrozak D.R., Price D.A., Melenhorst J.J., Hill B.J., Geldmacher C., Casazza J.P., Chattopadhyay P.K., Roederer M. Differential association of programmed death-1 and CD57 with ex vivo survival of CD8+ T cells in HIV infection. J. Immunol. 2009;183:1120–1132. doi: 10.4049/jimmunol.0900182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elahi S., Dinges W.L., Lejarcegui N., Laing K.J., Collier A.C., Koelle D.M., McElrath M.J., Horton H. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat. Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolte L., Gaardbo J.C., Skogstrand K., Ryder L.P., Ersbøll A.K., Nielsen S.D. Increased levels of regulatory T cells (Tregs) in human immunodeficiency virus-infected patients after 5 years of highly active anti-retroviral therapy may be due to increased thymic production of naive Tregs. Clin. Exp. Immunol. 2009;155:44–52. doi: 10.1111/j.1365-2249.2008.03803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maude S., Barrett D.M. Current status of chimeric antigen receptor therapy for haematological malignancies. Br. J. Haematol. 2016;172:11–22. doi: 10.1111/bjh.13792. [DOI] [PubMed] [Google Scholar]
- 27.Ghorashian S., Pule M., Amrolia P. CD19 chimeric antigen receptor T cell therapy for haematological malignancies. Br. J. Haematol. 2015;169:463–478. doi: 10.1111/bjh.13340. [DOI] [PubMed] [Google Scholar]
- 28.Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maus M.V., Grupp S.A., Porter D.L., June C.H. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123:2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deeks S.G., Wagner B., Anton P.A., Mitsuyasu R.T., Scadden D.T., Huang C., Macken C., Richman D.D., Christopherson C., June C.H. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol. Ther. 2002;5:788–797. doi: 10.1006/mthe.2002.0611. [DOI] [PubMed] [Google Scholar]
- 31.Mitsuyasu R.T., Anton P.A., Deeks S.G., Scadden D.T., Connick E., Downs M.T., Bakker A., Roberts M.R., June C.H., Jalali S. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4(+) and CD8(+) T cells in human immunodeficiency virus-infected subjects. Blood. 2000;96:785–793. [PubMed] [Google Scholar]
- 32.Scholler J., Brady T.L., Binder-Scholl G., Hwang W.T., Plesa G., Hege K.M., Vogel A.N., Kalos M., Riley J.L., Deeks S.G. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl. Med. 2012;4:132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., Patel B., Ghanem M.H., Bundoc V., Zheng Z., Morgan R.A., Rosenberg S.A., Dey B., Berger E.A. Novel CD4-based bispecific chimeric antigen receptor designed for enhanced anti-HIV potency and absence of HIV Entry receptor activity. J. Virol. 2015;89:6685–6694. doi: 10.1128/JVI.00474-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corti D., Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 35.Kwong P.D., Mascola J.R., Nabel G.J. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 36.Dragic T., Litwin V., Allaway G.P., Martin S.R., Huang Y., Nagashima K.A., Cayanan C., Maddon P.J., Koup R.A., Moore J.P., Paxton W.A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 37.Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R.E., Hill C.M. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y., Paxton W.A., Wolinsky S.M., Neumann A.U., Zhang L., He T., Kang S., Ceradini D., Jin Z., Yazdanbakhsh K. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 39.Allers K., Hütter G., Hofmann J., Loddenkemper C., Rieger K., Thiel E., Schneider T. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 40.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 41.Didigu C.A., Wilen C.B., Wang J., Duong J., Secreto A.J., Danet-Desnoyers G.A., Riley J.L., Gregory P.D., June C.H., Holmes M.C., Doms R.W. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61–69. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mock U., Machowicz R., Hauber I., Horn S., Abramowski P., Berdien B., Hauber J., Fehse B. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015;43:5560–5571. doi: 10.1093/nar/gkv469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romano Ibarra G.S., Paul B., Sather B.D., Younan P.M., Sommer K., Kowalski J.P., Hale M., Stoddard B., Jarjour J., Astrakhan A. Efficient modification of the CCR5 locus in primary human T cells with megaTAL nuclease establishes HIV-1 resistance. Mol. Ther. Nucleic Acids. 2016;5:e352. doi: 10.1038/mtna.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhoj V.G., Thibodeaux S.R., Levine B.L. Novel gene and cellular therapy approaches for treating HIV. Discov. Med. 2016;21:283–292. [PubMed] [Google Scholar]
- 47.Hubbard N., Hagin D., Sommer K., Song Y., Khan I., Clough C., Ochs H.D., Rawlings D.J., Scharenberg A.M., Torgerson T.R. Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood. 2016;127:2513–2522. doi: 10.1182/blood-2015-11-683235. [DOI] [PubMed] [Google Scholar]
- 48.Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porteus M.H. Towards a new era in medicine: therapeutic genome editing. Genome Biol. 2015;16:286. doi: 10.1186/s13059-015-0859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.P., Wang S.K., Ramos A., Chan-Hui P.Y., Moyle M., Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudicell R.S., Kwon Y.D., Ko S.Y., Pegu A., Louder M.K., Georgiev I.S., Wu X., Zhu J., Boyington J.C., Chen X., NISC Comparative Sequencing Program Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J., Ofek G., Yang Y., Zhang B., Louder M.K., Lu G., McKee K., Pancera M., Skinner J., Zhang Z., NISC Comparative Sequencing Program Mining the antibodyome for HIV-1-neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains. Proc. Natl. Acad. Sci. USA. 2013;110:6470–6475. doi: 10.1073/pnas.1219320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Z., Chinnasamy N., Morgan R.A. Protein L: a novel reagent for the detection of chimeric antigen receptor (CAR) expression by flow cytometry. J. Transl. Med. 2012;10:29. doi: 10.1186/1479-5876-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rooney C.M., Smith C.A., Ng C.Y., Loftin S.K., Sixbey J.W., Gan Y., Srivastava D.K., Bowman L.C., Krance R.A., Brenner M.K., Heslop H.E. Infusion of cytotoxic T cells for the prevention and treatment of Epstein-Barr virus-induced lymphoma in allogeneic transplant recipients. Blood. 1998;92:1549–1555. [PubMed] [Google Scholar]
- 57.Sommermeyer D., Hudecek M., Kosasih P.L., Gogishvili T., Maloney D.G., Turtle C.J., Riddell S.R. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali A., Kitchen S.G., Chen I.S., Ng H.L., Zack J.A., Yang O.O. HIV-1-Specific Chimeric Antigen Receptors Based on Broadly Neutralizing Antibodies. J. Virol. 2016;90:6999–7006. doi: 10.1128/JVI.00805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakrabarti B.K., Walker L.M., Guenaga J.F., Ghobbeh A., Poignard P., Burton D.R., Wyatt R.T. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J. Virol. 2011;85:8217–8226. doi: 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brudno J.N., Kochenderfer J.N. Toxicities of chimeric antigen receptor T cells: recognition and management. Blood. 2016;127:3321–3330. doi: 10.1182/blood-2016-04-703751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., Grupp S.A., Mackall C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hill A.L., Rosenbloom D.I., Goldstein E., Hanhauser E., Kuritzkes D.R., Siliciano R.F., Henrich T.J. Real-time predictions of reservoir size and rebound time during antiretroviral therapy interruption trials for HIV. PLoS Pathog. 2016;12:e1005535. doi: 10.1371/journal.ppat.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laird G.M., Bullen C.K., Rosenbloom D.I., Martin A.R., Hill A.L., Durand C.M., Siliciano J.D., Siliciano R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015;125:1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milone M.C., Fish J.D., Carpenito C., Carroll R.G., Binder G.K., Teachey D., Samanta M., Lakhal M., Gloss B., Danet-Desnoyers G. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 2009;17:1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Challita P.M., Skelton D., el-Khoueiry A., Yu X.J., Weinberg K., Kohn D.B. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clouse K.A., Powell D., Washington I., Poli G., Strebel K., Farrar W., Barstad P., Kovacs J., Fauci A.S., Folks T.M. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J. Immunol. 1989;142:431–438. [PubMed] [Google Scholar]
- 67.Folks T.M., Clouse K.A., Justement J., Rabson A., Duh E., Kehrl J.H., Fauci A.S. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. USA. 1989;86:2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buttke T.M., Folks T.M. Complete replacement of membrane cholesterol with 4,4′,14-trimethyl sterols in a human T cell line defective in lanosterol demethylation. J. Biol. Chem. 1992;267:8819–8826. [PubMed] [Google Scholar]
- 69.Folks T., Benn S., Rabson A., Theodore T., Hoggan M.D., Martin M., Lightfoote M., Sell K. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc. Natl. Acad. Sci. USA. 1985;82:4539–4543. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J., Press O.W., Lindgren C.G., Greenberg P., Riddell S., Qian X., Laugen C., Raubitschek A., Forman S.J., Jensen M.C. Cellular immunotherapy for follicular lymphoma using genetically modified CD20-specific CD8+ cytotoxic T lymphocytes. Mol. Ther. 2004;9:577–586. doi: 10.1016/j.ymthe.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 71.Cann A.J., Zack J.A., Go A.S., Arrigo S.J., Koyanagi Y., Green P.L., Koyanagi Y., Pang S., Chen I.S. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 1990;64:4735–4742. doi: 10.1128/jvi.64.10.4735-4742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koyanagi Y., Miles S., Mitsuyasu R.T., Merrill J.E., Vinters H.V., Chen I.S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 73.McClure J., van’t Wout A.B., Tran T., Mittler J.E. Granulocyte-monocyte colony-stimulating factor upregulates HIV-1 replication in monocyte-derived macrophages cultured at low density. J. Acquir. Immune Defic. Syndr. 2007;44:254–261. doi: 10.1097/QAI.0b013e318030f5c5. [DOI] [PubMed] [Google Scholar]
- 74.Sourisseau M., Sol-Foulon N., Porrot F., Blanchet F., Schwartz O. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 2007;81:1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.