Figure 2.
CRISPR Activity in Myoblasts with Normal-Size (CTG⋅CAG)n Repeats
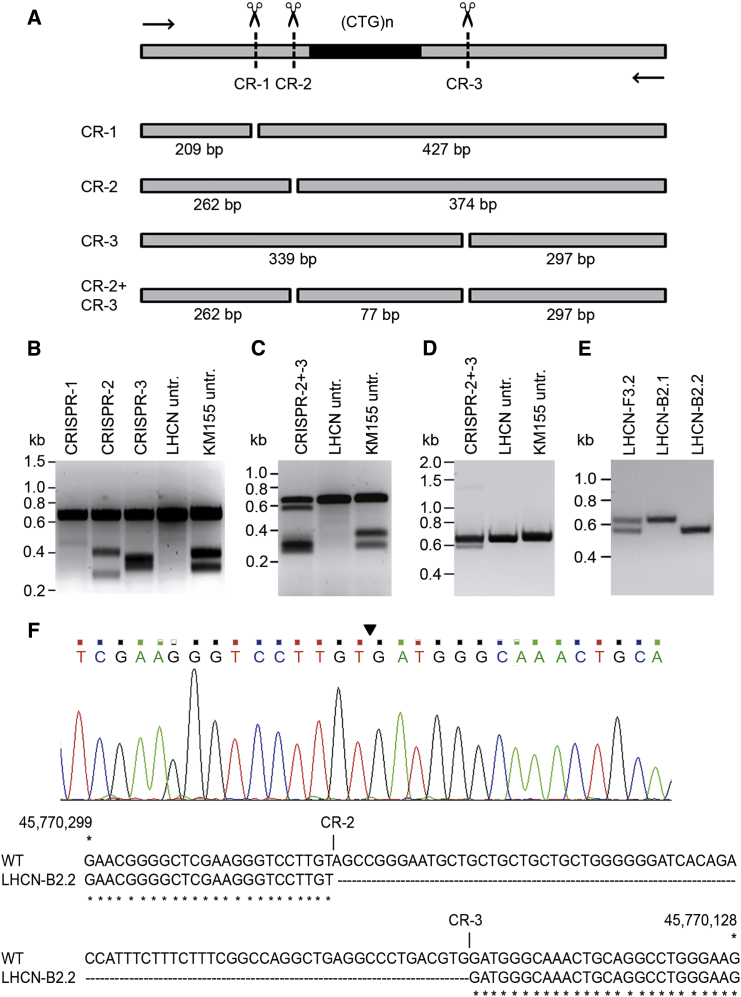
(A) Schematic outline of the T7EI assay for determination of CRISPR cleavage efficiency. Part of DMPK exon 15 ([CTG⋅CAG]n repeat in black) containing CRISPR-1, -2, and -3 recognition sites and positioning of PCR primers used for amplification of the relevant segment are shown on top. Possible fragments formed in the assay are depicted, with sizes given underneath. (B) T7EI assay of DNA from small pools of transfection-positive LHCN cells. Quantification of signal strength (assessed by scanning of fluoresce signal intensity upon UV illumination for all assays shown in B–E) revealed target efficiencies of <1% for CRISPR-1, 8%–21% for CRISPR-2, and 14% for CRISPR-3. DNAs of non-transfected (untr.) LHCN (two alleles with equal DMPK repeat lengths) and KM155C25 myoblasts (one [CTG⋅CAG]5 and one [CTG⋅CAG]14 allele) were used as negative and positive control, respectively. (C) T7E1 assay of DNA from a pool of LHCN myoblasts treated with CRISPR-2 and -3 simultaneously. Untransfected LHCN and KM155C25 were included as controls. Note that we could not differentiate between deletion of the entire region between the two CRISPR sites or simultaneous formation of small indels at each of the two CRISPR sites. (D) PCR analysis of the relevant DMPK genomic segment after dual genome-editing with CRISPR-2 and -3. The upper band represents the unmodified PCR product. The lower band is indicative for deletion of the 77 bp (CTG⋅CAG)5 repeat and flanking regions in a small portion of CRISPR-2- and CRISPR-3-treated cells. (E) PCR analysis of genome changes in three CRISPR-2- and CRISPR-3-treated LHCN cell clones. Clone LHCN-B2.1 contains two unmodified repeats (single signal at 636 bp), whereas clone LHCN-B2.2 has a repeat deletion on both alleles (single signal at 559 bp). Clone LHCN-F3.2 carries one unmodified and one edited allele (signals at 559 bp and 636 bp). (F) Sequence verification of excision of the repeat-containing segment. Top: sequencing profile of the DMPK exon-15 gene region in clonally expanded LHCN cells after dual gene editing with CRISPR-2 and -3. The site at which the DSBs are fused is indicated by an arrowhead. No indels were found. Bottom: the exon-15 sequence lacking the 77-bp repeat-containing segment aligned with the normal DMPK sequence.