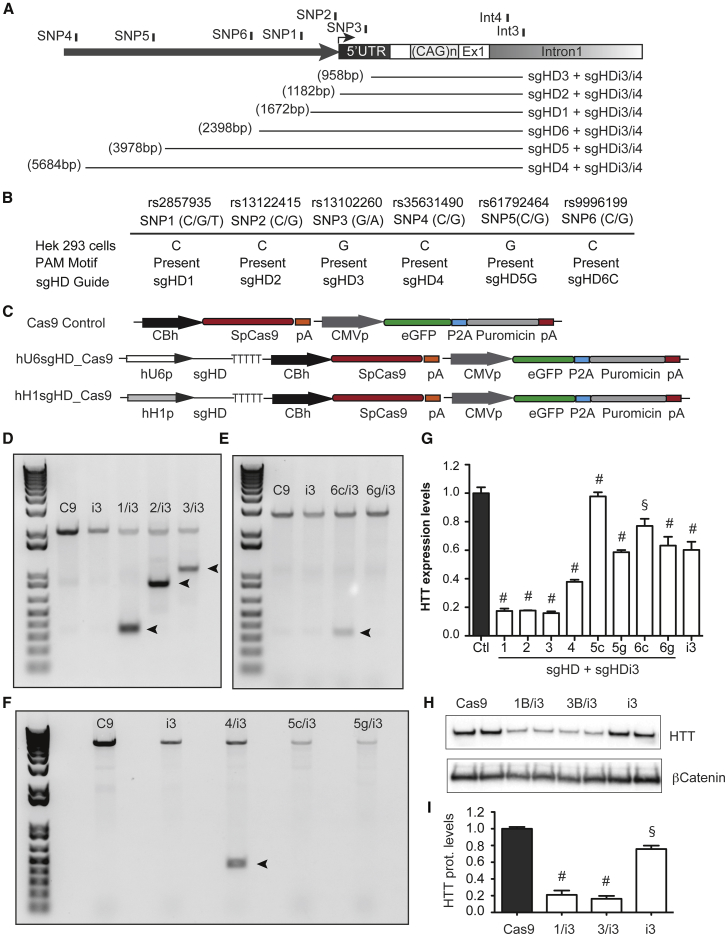
Figure 2.
Cleavage of SNP-Dependent sgHD/SpCas9 Complexes in HEK293 Cells
(A) Cartoon depicting the relative position of the six prevalent SNP-dependent PAMs upstream of HTT exon-1 and two common PAMs within HTT intron-1. The estimated size of the targeted deleted sequence is indicated. (B) The genotype of the prevalent SNPs within the HTT promoter in HEK293 cells is shown. All SNPs were homozygous for the nucleotide variation and the PAM motif was present for the sgRNA indicated. (C) A diagram of the CRISPR expression systems transfected into HEK293 cells is shown. (D–F) A genomic PCR showing HTT exon-1-targeted deletion by sgRNA/SpCas9 pair complexes binding upstream and downstream of the target sequence is shown in the images. (G) RT-qPCR analysis of HTT mRNA levels in HEK293 cells transfected with sgHD/SpCas9 expression cassettes targeting upstream promoter SNPs and the common intronic sgHDi3 sequence is shown. All of the samples are normalized to human GAPDH, and the results are the mean ± SEM relative to cells transfected with plasmids containing the SpCas9 only control (n = 6 independent experiments; §p < 0.001, #p < 0.0001, and one-way ANOVA followed by a Bonferroni’s post hoc). (H) sgHD1/i3/SpCas9, sgHD3/i3/SpCas9, and sgHDi3/SpCas9 expression cassettes were transfected into HEK293 cells, and endogenous HTT protein levels were determined after puromycin selection and expansion. Cells transfected with Cas9 only were used as a control and beta catenin served as a loading control. (I) The quantification of HTT protein levels after treatment with sgHD/SpCas9 complexes is shown. The data are the mean ± SEM relative to cells transfected with plasmids containing SpCas9 only control (n = 6 independent experiments; #p < 0.0001, §p < 0.001, and one-way ANOVA followed by Bonferroni’s post hoc).