Abstract
Duchenne muscular dystrophy (DMD) is a lethal, X-linked, muscle-wasting disorder caused by mutations in the large, 2.4-Mb dystrophin gene. The majority of DMD-causing mutations are sporadic, multi-exon, frameshifting deletions, with the potential for variable immunological tolerance to the dystrophin protein from patient to patient. While systemic gene therapy holds promise in the treatment of DMD, immune responses to vectors and transgenes must first be rigorously evaluated in informative preclinical models to ensure patient safety. A widely used canine model for DMD, golden retriever muscular dystrophy, expresses detectable amounts of near full-length dystrophin due to alternative splicing around an intronic point mutation, thereby confounding the interpretation of immune responses to dystrophin-derived gene therapies. Here we characterize a naturally occurring deletion in a dystrophin-null canine, the German shorthaired pointer. The deletion spans 5.6 Mb of the X chromosome and encompasses all coding exons of the DMD and TMEM47 genes. The sequences surrounding the deletion breakpoints are virtually identical, suggesting that the deletion occurred through a homologous recombination event. Interestingly, the deletion breakpoints are within loci that are syntenically conserved among mammals, yet the high homology among this subset of ferritin-like loci is unique to the canine genome, suggesting lineage-specific concerted evolution of these atypical sequence elements.
Keywords: Duchenne muscular dystrophy, concerted evolution, pseudogene, dystrophin-null, German shorthaired pointer
Introduction
Recent progress in vector-mediated gene therapy shows promise in the treatment of Duchenne muscular dystrophy (DMD).1 However, as in the case of many genetic diseases, a protein is mutated or altogether absent, preventing the establishment of immunological tolerance to its wild-type form. Thus, gene therapies that deliver a transgene modeled after a wild-type protein may contain epitopes to which the patient’s immune system lacks central tolerance, and, therefore, they risk inciting a deleterious host immune response.2, 3
In addition, the sporadic and highly varied dystrophin mutations within the DMD patient population make evaluation of immune responses following treatment exceptionally challenging, as each patient’s immune system may react differently to the peptide product of a recombinant transgene. Therefore, an animal model void of immunological tolerance to all dystrophin epitopes should provide the most sensitive prediction of immune responses to gene therapies.
Preclinical development of gene therapies for DMD has centered on the use of two naturally occurring animal models, the mdx mouse and the golden retriever muscular dystrophy (GRMD) dog, which are caused, respectively, by a nonsense mutation within exon 234 and a point mutation within the splice acceptor site of intron 6.5 These mutations represent only a small portion of those seen in the DMD patient population; therefore, they cannot accurately predict the potential human immune responses to DMD gene therapies, and there are currently no primate models for DMD. Furthermore, naturally occurring exon skipping and stop codon readthrough can result in leaky dystrophin expression, as evidenced by revertant fibers,6, 7, 8 and they may allow for the establishment of immunological tolerance to dystrophin during development in these animal models. Alternatively, dystrophin expression in revertant fibers could result in a primed immune response to dystrophin peptides, as was shown in humans.9 Both of these outcomes convolute the interpretation of immune responses to newly produced proteins acting as neoantigens.
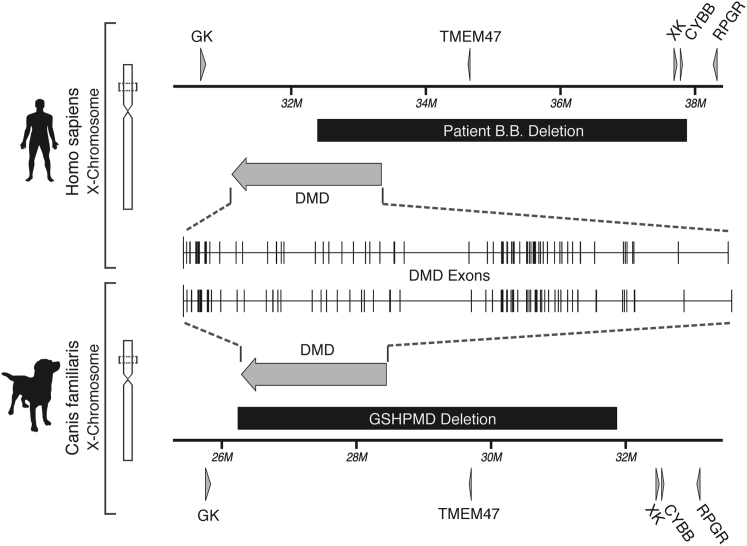
The German shorthaired pointer-muscular dystrophy (GSHPMD) is a recently described, naturally occurring dog model of DMD.10 Western blot and fluorescence in situ hybridization (FISH) analyses suggest that the deletion in this model may encompass the entirety of the dystrophin gene.11 Here we used a PCR approach to precisely define the deletion endpoints and to confirm the complete absence of the DMD gene by sequencing across the deletion. We found that the deletion spans 5.6 Mb and is remarkably similar to that of patient B.B., a boy whose deletion dramatically accelerated the characterization of the DMD locus (Figure 1).12, 13, 14, 15, 16 Interestingly, the GSHPMD deletion breakpoints are within highly homologous DNA loci that are conserved on the mammalian X chromosome. The GSHPMD model, lacking the entirety of the dystrophin gene and, therefore, void of any possible level of immunological tolerance or sensitivity to wild-type dystrophin epitopes, could provide a much-needed platform for the prediction of immune responses to gene therapies for DMD.
Figure 1.
Comparison of Human and Dog DMD and Neighboring Genes
Orthologous regions of the human and dog X chromosome surrounding the DMD gene are compared. Arrows indicate genes, with the DMD gene expanded inward for comparison of exons between the two species. The X chromosome deletion in the historic patient, B.B., is depicted in a black box, as is the GSHPMD deletion. Arrowheads indicate the direction of transcription. GK, glycerol kinase; DMD, dystrophin; TMEM47, transmembrane protein 47; XK, X-linked Kx blood group; CYBB, cytochrome b-245 beta polypeptide; RPGR, retinitis pigmentosa GTPase regulator; M, million.
Results
Despite 100 million years of evolution,17 the regions of the human and canine X chromosomes that encompass DMD, including exon-intron spacing and size, are conserved (Figure 1), perhaps indicative of the vital role its encoded peptide, dystrophin, plays in muscle biology. For clarity in the presentation of our results, we reference the default orientation and numbering of the X chromosome delegated by the NCBI, which assigns DMD to the antisense strand in the dog genome assembly (Figure 1).
Genetic Mapping of the GSHPMD Deletion Breakpoints
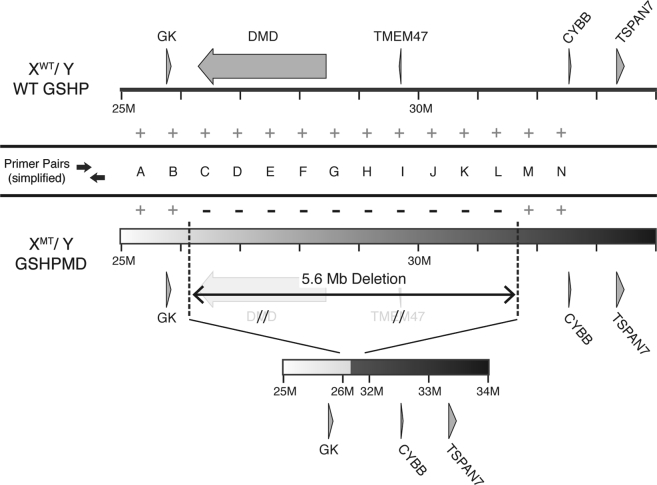
To map the deletion in the GSHPMD model, we designed a PCR-based strategy to locate the breakpoints on the X chromosome. Using previously reported FISH data from a GSHPMD carrier female,11 we estimated the location of the deletion and designed primer pairs that broadly spanned this region of the X chromosome (Figure 2). Gel electrophoresis was used to compare PCR results from wild-type (WT) dog or GSHPMD DNA to broadly map the deleted region of the X chromosome. Following a similar approach, additional primer pairs were designed to finely map the telomeric (TBP) and centromeric (CBP) breakpoints. In this way, we employed 74 unique primer pairs and mapped the TBP to be within base pairs 26,237,921–26,239,551 and the CBP to be within base pairs 31,867,082–31,871,007 of the dog X chromosome, proving that the deletion spans 5.6 Mb and encompasses the entirety of the DMD and TMEM47 genes.
Figure 2.
Deletion Map of the GSHPMD Model by PCR
The X chromosome of male wild-type GSHP and affected GSHPMD dogs are compared. The locations of primer pairs are labeled alphabetically. The results of each PCR experiments are shown, with plus indicating successful amplification and minus indicating failed amplification. The mapped deletion in the GSHPMD model is depicted inferiorly. XWT/Y, wild-type male; XMT/Y, mutant male; PCR, polymerase chain reaction; TSPAN7, tetraspan 7; Mb, million base pairs; M, million.
Further attempts to more finely map the deletion breakpoints repeatedly failed (data not shown), perhaps due to a 711-bp genome assembly gap at base pairs 26,238,732–26,239,442 within the TBP region (Figure S1A). To determine the DNA sequence of the assembly gap, we used primer pair 74 to PCR-amplify the respective genomic region from a BAC clone from the library used in the dog genome assembly project (Figure S1B).18 Amplification of this region was difficult and dependent on the addition of betaine, a PCR enhancer, which may explain why the region was not sequenced in the dog genome assembly. Following amplification, we gel-purified and sequenced the 1.5-kb PCR product and assembled the individual Sanger reads into a single DNA contig (GenBank: KR907258; Figure S1C). A Pustell DNA matrix was used to compare the assembled contig to the region of the X chromosome that harbors the assembly gap (Figures S1D and S6). This comparison revealed >97% homology between our sequenced PCR product and the chromosomal sequence flanking the genome assembly gap, suggesting that the internal 650-bp region of the PCR product sequence is representative of the previously unknown assembly gap sequence. We therefore replaced base pairs 26,238,718–26,239,484 of the dog X chromosome with base pairs 338–1,045 of our BAC-derived, PCR product, and we used the resulting sequence in our subsequent analyses.
The GSHPMD Deletion Spans 5.6 Mb and Is Contiguous
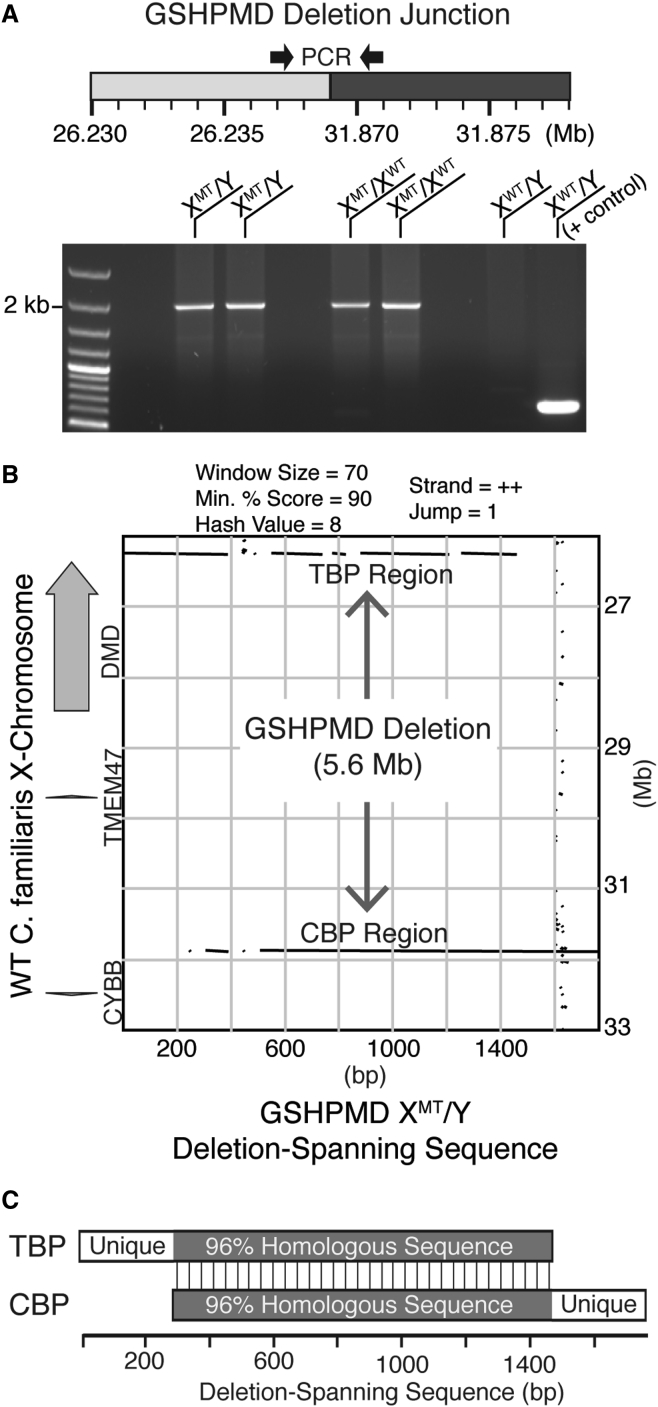
To confirm the absence of the entire 5.6-Mb DNA fragment from the X chromosome, we designed a primer pair that flanks the predicted deletion. The primer pair sequences encompass 5.6 Mb, which is far outside the limits of PCR. However, unique to PCRs containing DNA from affected GSHPMD males and carrier females, a 2-kb amplicon was generated, suggesting that the entire 5.6-Mb region is deleted from the X chromosome in the GSHPMD model (Figure 3A).
Figure 3.
PCR Amplification across the 5.6-Mb Deletion in the GSHPMD Model
(A) Schematic showing primers that are spaced over 5.6 Mb apart in wild-type dog. PCR products generated from this primer pair using DNA from two affected males, two carrier females, and one wild-type male are displayed on an agarose gel following electrophoresis. A positive control using an unrelated primer pair is provided in the rightmost lane for the wild-type dog. (B) Pustell DNA matrix comparing the sequenced deletion-spanning amplicon from a GSHPMD male to the indicated region of wild-type dog X chromosome. Black lines indicate homology between the compared sequences. (C) Macroscale DNA sequence comparison of the telomeric and centromeric deletion breakpoints. Shaded region indicates >96% homology between the deletion breakpoints. XMT/Y, affected male; XMT/XWT, carrier female; XWT/Y, wild-type male; TBP, telomeric breakpoint; CBP, centromeric breakpoint; WT, wild-type.
To confirm its identity, we gel-purified and sequenced the GSHPMD-specific amplicon, and subsequently we assembled the individual Sanger reads into a 1.7-kb contig (GenBank: KR907259). A Pustell DNA matrix comparing the deletion-spanning contig and base pairs 26–33 Mb of the dog X chromosome revealed >90% sequence homology between the 5′ portion of the contig and the TBP region, with an eventual, subtle shift in homology to favor the CBP region (Figure 3B), consistent with the sequenced amplicon-spanning deletion. A short break in homology near the TBP was present in our original Pustell matrix, but it was found to result from three insertions or deletions (indels) not accommodated by the matrix (Figure S2A). In addition, a break in homology attributable to a TAAA tandem repeat was present between 400 and 500 bp of the Pustell matrix. The abundance of this repeat became apparent when specificity parameters of the Pustell matrix were reduced (Figure S2B).
To our surprise, our Pustell matrix (Figure 3B) also revealed that the TBP and CBP regions were highly homologous to each other, raising questions as to the mechanism of the deletion. Importantly, both the 5′ and 3′ ends of the deletion-spanning contig extended into regions unique to the TBP and CBP, respectively, confirming that, despite the homology surrounding both breakpoints, the deletion-spanning contig indeed spans the GSHPMD deletion (Figure 3C). Taken together, these findings demonstrate that the entirety of the mapped 5.6-Mb region, encompassing the DMD and TMEM47 genes, is deleted from the X chromosome in the GSHPMD model and that the deletion breakpoints are highly homologous to each other.
Identification of Homologous FTHL Loci Present on the Dog X Chromosome
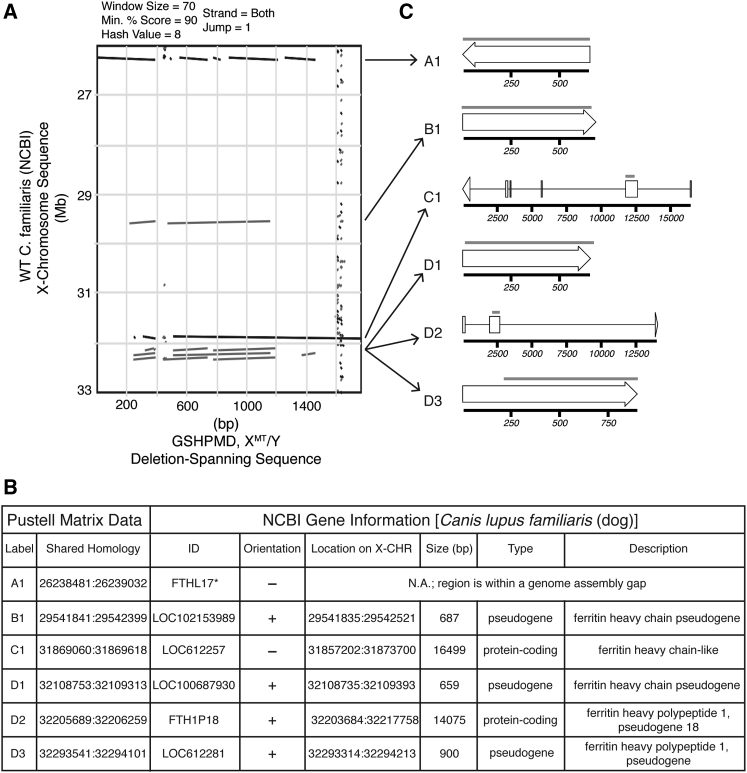
To further investigate the identified homology between the deletion breakpoints, we generated a Pustell matrix comparing the DNA sequence of the deletion-spanning contig and 26- to 33-Mb base pairs of the dog X chromosome, allowing for comparison of DNA in both orientations. This revealed four additional locations of the X chromosome with >90% homology to the deletion-spanning contig, although in opposite orientation to the previously identified deletion breakpoints (Figure 4A).
Figure 4.
Identification of Homologous DNA Segments on Dog X Chromosome as Members of the Ferritin-like Superfamily
(A) Pustell DNA matrix comparing both orientations of the sequenced deletion-spanning amplicon from a GSHPMD male to the indicated region of wild-type dog X chromosome from the dog reference genome. Strand homology is provided in the subsequent table. (B) Regions of homology identified in Pustell matrix and corresponding gene annotations for these regions of the dog genome from NCBI. Loci are grouped and labeled based on chromosomal location. (C) Schematic of identified ferritin-like genes and pseudogenes. Arrows indicate the location of the genes and pseudogenes in the Pustell matrix. Area of shared homology is shown with a gray bar above each respective locus. *Gene inferred from mammalian FTHL17 ortholog.
To determine the identity of the six homologous regions, we approximated their locations on the X chromosome and searched for available annotations within the dog genome assembly via NCBI. Five of the six homologous regions were annotated in the dog genome, and, surprisingly, all five regions were designated as ferritin heavy chain-like (FTHL), two being protein-coding genes and three being pseudogenes (Figure 4B). Importantly, the unidentified, sixth region overlapped with the TBP and extended into the aforementioned assembly gap, likely accounting for its lack of annotation in the dog genome assembly. We found that, while the shared homology spanned nearly the entire length of the pseudogenes, it was unique to only a portion of the protein-coding genes, specifically exon 2 (Figure 4C). Using the encoded peptide sequence of exon 2 of these genes, we queried reference proteomes using HMMER,19 which uses hidden Markov modeling to search for homologous proteins, and we found that exon 2 is similar to the ferritin-like domain (Data S1). Intriguingly, a DNA sequence alignment of the six homologous regions identified in our Pustell matrix revealed that >96% identities are conserved between the pseudogenes and exon 2 of the protein-coding genes (Figure S7), raising questions as to the mechanism responsible for such high sequence conservation among FTHL genes and pseudogenes alike.
Of the identified ferritin loci, two have NCBI descriptions that link them to the protein-coding, ferritin heavy polypeptide 1 gene (FTH1) (Figure 4B). Of note, we identified two copies of FTH1 in the dog, one with introns located on chromosome 18 and another lacking introns located on chromosome 11 (Figure S3A), and we demonstrated that the cDNA sequences of the dog FTH1 genes are 100% identical. Further examination revealed a polyadenylation (polyA) signal, AATAAA, followed closely by a string of forty adenine residues, both well-characterized hallmarks of processed mRNAs, uniquely in the intron-lacking copy of the dog FTH1 gene on chromosome 11 (Figure S3).20, 21, 22 A brief search of NCBI suggested that the additional, intron-lacking copy of FTH1 is unique to the dog, despite the fact that the intron-containing gene is evolutionarily ancient, as marked by the presence of an ortholog in a wide range of species, including the elephant shark, chicken, cat, and human (data not shown). These findings provide strong evidence to suggest that the intron-lacking copy of FTH1 on dog chromosome 11 is a processed pseudogene, known to arise through reverse transcription and integration of an mRNA, and, further, that it is of recent origin.
Next, we questioned whether the identified FTHL pseudogenes (Figure 4B) might actually be processed pseudogenes. We generated a ClustalW sequence alignment of the six homologous regions, but this time we extended the lengths of the individual sequences in the 3′ direction. To our surprise, we identified a polyA signal followed by an A-rich stretch of DNA that varied in length in all six homologous regions, perhaps representing the remnants of what was once a polyA tail but that also could be an unrelated tandem repeat (Figure S2B). We included the second exons and a portion of their 3′ introns from the two protein-coding genes, LOC612257 and FTH1P18, in our alignment, and we found that the polyA signal and A-rich stretch are located just inside the 3′ intron (Figures S3D and S8). While the dog FTH1 processed pseudogene contains a 100% intact polyA signal and tail suggesting it is of recent origin, the identified FTHL genes and pseudogenes contain a less indicative polyA tail, more in line with an earlier origin in evolutionary history, and, therefore, also might be present in other species.
The Identified FTHL Loci Pseudogenes Are Syntenically Conserved among Mammals
The ferritins are evolutionarily ancient iron-binding proteins that are part of the large ferritin-like superfamily. Many copies of the ferritin-H subunit are known to exist as processed pseudogenes in the human.23, 24, 25 Therefore, we asked whether the identified dog FTHL loci (Figure 4B) also are present in other mammals.
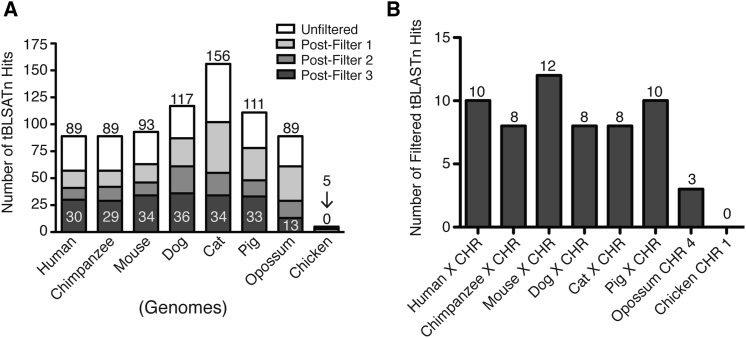
To probe this possibility, we used a phylogenetic approach to determine when in evolutionary history the FTHL loci originated. Using the dog FTH1 amino acid sequence as query, we performed a tBLASTn search26 of mammalian (human, chimpanzee, mouse, dog, cat, and pig), marsupial (opossum), and aves (chicken) genomes. Large numbers of hits were returned for all species, with the exception of the chicken (Figure 5A). To select for true FTHL-processed pseudogenes and intron-lacking genes, we established criteria by which to filter the tBLASTn results (Figure S4). In brief, filtering tBLASTn hits by length and identity criteria substantially reduced the number of hits returned by our search. Interestingly, a comparison of the filtered tBLASTn hits located on the DMD-containing chromosome of the queried species revealed that mammals contained a relatively similar numbers of hits, whereas marsupials contained fewer, and aves contained none (Figure 5B).
Figure 5.
Quantification of Results from tBLASTn Search of Dog FTH1 Peptide in Multiple Species
(A) Bar graph quantifying tBLASTn search results of dog FTH1 peptide sequence against the genomes of the indicated species. Number at the top of each bar signifies hits returned prior to applying filtering criteria. Number internal to bottom section of each bar represents hits remaining after applying all filtering criteria. (B) Bar graph quantifies the number of filtered tBLASTn hits present on the chromosome that harbors the DMD gene for the indicated species. CHR, chromosome.
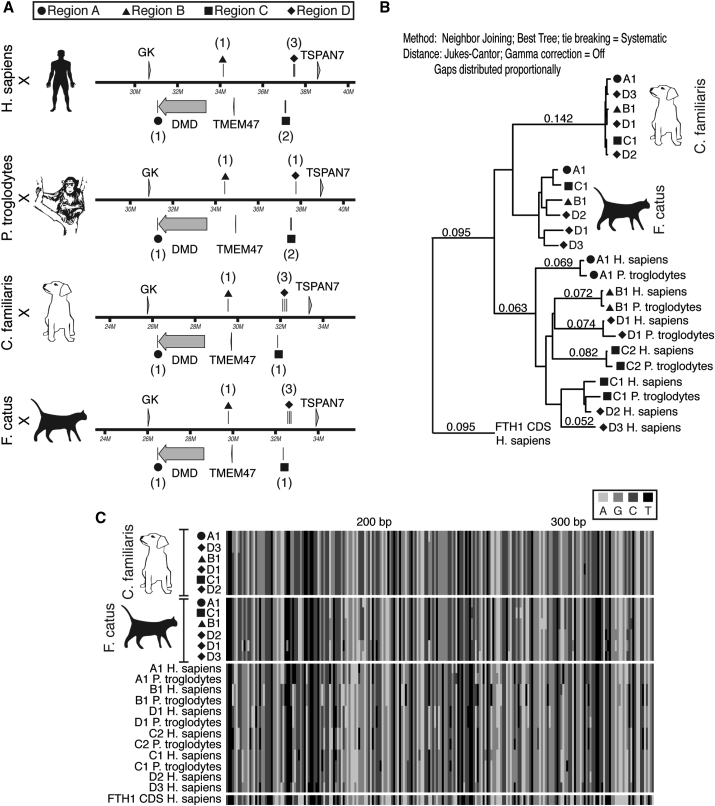
In addition, a syntenic, multi-species comparison of DMD and its surrounding chromosomal region revealed that the locations of the earlier identified dog FTHL loci (Figure 4B) are conserved in mammals, and they can be grouped into four chromosomal regions (Figures 6A and S5A). Although the number of FTHL loci within each of the syntenic chromosomal regions varied by species, our tBLASTn search revealed at least one copy per region in the queried mammals, with the exception of the mouse and the four corresponding syntenic regions of the opossum and chicken (Table S1). In agreement with our tBLASTn search, NCBI designated nearly all of the mammalian hits as FTHL genes or pseudogenes. While not yet formally annotated in the dog genome, our phylogenetic analysis shows that the unidentified dog locus, located in the TBP of the GSHPMD deletion, is sytenically conserved among several mammalian species, suggesting that it is the dog ortholog of the human ferritin heavy polypeptide-like 17 (FTHL17) gene. Furthermore, a multi-species sequence alignment and phylogeny of the FTHL genes and pseudogenes revealed that, of the species examined, the extreme homology among the identified X chromosome FTHL loci is unique to the dog. Interestingly, a subset of the FTHL loci also was highly homologous in the mouse, though this subset was closely linked, spanning less than 85 kb, as compared to the homology shared among the entire set of the identified FTHL loci that span more than 6 Mb in the dog (Figures 6B and S5).
Figure 6.
Phylogenetic Analysis of Identified FTHL Loci from tBLASTn Search in Human, Chimpanzee, Dog, and Cat
(A) Comparison of syntenic portion of X chromosome from each species showing the grouping of FTHL loci to four regions, labeled A, B, C, and D. Number of pseudogenes present in reach region is indicated in parentheses. (B) Phylogeny of identified FTHL loci. The human FTH1 CDS is used as an outgroup. Branches with values less than 0.05 are not displayed. Tree-building parameters are provided as text in figure. CDS, coding DNA sequence. (C) ClustalW multiple DNA sequence alignment of identified FTHL loci. Note extreme sequence homology among dog FTHL loci.
In summary, these findings suggest that the identified FTHL genes and pseudogenes are evolutionarily ancient, arising prior to mammalian radiation. Yet, despite millions of years of evolution, the DNA sequences of these FTHL genes and pseudogenes in the dog, and to a lesser degree the mouse, have remained highly homologous. However, pseudogenes, if free from selective pressure, would be expected to diverge in DNA sequence over time, as is seen in the human and chimpanzee (Figure 6). This intriguing finding suggests a possible function, perhaps at the DNA level, for the identified canine FTHL pseudogenes.
Discussion
In this study, we show that the deletion in the GSHPMD model spans 5.6 Mb of the canine X chromosome and encompasses all known exons of the contiguous DMD and TMEM47 genes. Importantly, the GSHPMD model is expected to be devoid of immunological tolerance and sensitization to dystrophin, due to the complete deletion of its respective gene, and, therefore, it should provide an instrumental pre-clinical model for the prediction of immune responses to gene therapies for DMD. Sequence confirmation of the deletion breakpoint insures that no coding exons of DMD remain, precluding expression of any dystrophin-derived peptide sequences through exon skipping or alternative transcription from any known internal promoters. The same considerations apply to the contiguous TMEM47 gene, which encodes a highly conserved 19.9-kDa protein without any identified Mendelian disease association. TMEM47, also known as brain cell membrane protein I, is abundantly transcribed in dog brain.27 To the best of our knowledge, TMEM47 protein expression has not been characterized in the dog; we were unable to detect TMEM47 protein in brain tissue of wild-type dog by immunohistochemistry or western blot, using a commercially available polyclonal antibody raised against a shared human epitope, and there are no canine-reactive antibodies commercially available (data not shown). TMEM47’s candidacy for a role in human X-linked mental retardation has been explored and dismissed.28 Moreover, the phenotypic difference between dystrophic and normal littermates in the GSHPMD colony appears to be strictly related to muscle impairment, as is the case in the GRMD colony.
The deletion breakpoints were within highly homologous segments of DNA that NCBI identifies as members of the ferritin family. We uncovered additional copies of the homologous ferritin DNA loci on the dog X chromosome, designated as a mix of protein-coding genes and pseudogenes. These ferritin-like loci are evolutionarily ancient, arising prior to mammalian radiation; but, interestingly, the high homology is unique to the ferritin loci in the dog and, to a lesser degree, the mouse. The observed sequence homology among the identified ferritin genes and pseudogenes is maintained across large genetic distances (>6 Mb) uniquely in the dog, a phenomenon likely arising through gene conversion (reviewed in Chen et al.29). These findings suggest that the GSHPMD deletion occurred through a homologous recombination event, an outcome that was likely enabled by gene conversion of these ferritin loci in the canine lineage.
To map the deletion in the GSHPMD model, we used a PCR strategy and found no amplification from primers internal to the deletion. Furthermore, amplification across the 5.6-Mb deletion generated an amplicon uniquely in the GSHPMD model (Figures 2 and 3A), the length and sequence of which demonstrate a clean breakpoint without insertion or rearrangement. This, in combination with data showing that skeletal muscle of GSHPMD dogs lacks dystrophin,11 provides clear evidence supporting the deletion, rather than translocation, of the 5.6-Mb region of X chromosome that encompasses the DMD and TMEM47 genes. The deletion breakpoints are within highly homologous segments of DNA, which span over 1 kb and share 96% identity (Figure 3C), suggesting that the deletion occurred through a homologous recombination event. Supporting this claim, the region encompassing the TBP of the GSHPMD deletion is a recombination hotspot in the dog genome.30 Consistent with recombination hotspots in the dog,30 the GSHPMD deletion breakpoints have >67% GC content over a length of about 550 bp (Figure S7). Furthermore, the FTHL17 gene, which overlaps with the TBP of the GSHPMD model, has been implicated in the X chromosome translocation breakpoint of an infertile human male.31 Together, this indirect evidence strongly supports the notion that the GSHPMD deletion likely occurred through homologous recombination.
Next, we discovered that the deletion breakpoints were not only homologous to each other but also homologous to four additional regions of the dog X chromosome. These six regions share >96% identity over a length of about 550 bp, are distributed across 6 Mb of the X chromosome, and are members of the ferritin family (Figures 4 and S7). Interestingly, NCBI designates these ferritin loci as a mixture of protein-coding genes and pseudogenes. However, the high level of homology between the FTHL loci in the dog and a subset in the mouse suggests selective pressure on these loci, perhaps at the DNA level (Figures 6 and S5). Of note, a comparison of the FTHL hits returned by tBLASTn in the human and chimpanzee, which shared a common ancestor less than 10 million years ago,17 shows that interspecies orthologs are more closely related than intraspecies paralogs (Figure 6B), the expected outcome of divergent evolution. In contrast, the identified FTHL loci in the dog have not diverged and appear to be evolving in concert.
Concerted evolution describes a phenomenon in which individual genetic copies of a multigene family, generally present in a tandem array, evolve in concert; that is, a point mutation in one copy becomes propagated throughout the array of genes (reviewed in Nei and Rooney32). This phenomenon was first described during a study of the rRNA genes in the African toad33 and is thought to occur by gene conversion,29 a mechanism that utilizes DNA repair machinery to transfer base-pair mismatches between highly homologous yet distinct DNA strands following a double-stranded break. However, in contrast to the ∼450 repeat copies in rRNA genes of the African toad, we found only six highly homologous copies of the FTHL loci that span over 6 Mb on the canine X chromosome. Unequal crossover of the FTHL17 gene with any of the other identified FTHL loci would result in a large DMD-encompassing deletion, such as that seen in the GSHPMD model. However, given that the DMD phenotype is not commonly reported in dogs, the more recently proposed synthesis-dependent strand annealing (SDSA) model,34 which results in gene conversion yet non-crossover products, is a more plausible mechanism to account for the concerted evolution of the identified FTHL loci. Gene conversion is known to occur across long genetic distances in humans, as with von Willebrand disease, where mutation in the VWD gene can result from interchromosomal gene conversion between the true VWD gene and a highly homologous pseudogene.35
While arguing against the intrinsic definition of a pseudogene, we thought the identified FTHL pseudogenes might perform a function in the dog and would, therefore, be conserved due to selective pressure. Indeed, while most pseudogenes lose their transcription potential, those that are transcribed are capable of acting as short interfering RNAs and microRNAs.36 But, hindering our analysis, the FTHL17 gene has testis-specific activity in both the human and mouse,37, 38 although the human FTHL17 protein is unstable and has no ferroxidase activity.39 Perhaps unsurprisingly, a BLASTn40 search of the dog transcriptome41 using a FTHL sequence as query resulted in greater than 1,000 hits in testis and less than 10 hits in all other tissue types that were available (data not shown). However, due to the short, 100-bp sequence reads of the dog transcriptome and the extreme homology between the FTHL loci, we were not able to discern whether the observed hits were transcripts resulting from the FTHL genes or the FTHL pseudogenes. Dedicated study of canine FTHL pseudogenes may reveal a function as an RNA species.
Here we provide a detailed characterization of the DMD deletion in the GSHPMD model. Our data strongly suggest the deletion arose through homologous recombination of FTHL loci that appear to be evolving in concert in the dog. Due to the complete deletion of the DMD gene, this model is expected to be devoid of central immunological tolerance to any portion of the 427-kDa dystrophin protein. Furthermore, this deletion precludes any sensitization to truncated dystrophin peptides that might, with other mutations, arise somatically within revertant fibers. These features of the GSHPMD model favor its use in the rigorous study of cytotoxic immune responses to recombinant dystrophin expressed following regional or systemic gene therapy. Interestingly, intra-breed allelic diversity, while limited, has been identified within DLA-88, a dog leukocyte antigen (DLA) gene that displays the highest polymorphism of the MHC class I loci.42, 43, 44 Therefore, immunological studies within a single dog breed, such as the GSHPMD model, may result in varied responses based on DLA class I haplotype, a result that can be expected in clinical trials due to the highly polymorphic HLAs encoding MHC I.45 In conclusion, we envision the GSHPMD model will be instrumental in the evaluation of potential immune responses to gene therapies for DMD, which will aid in the demonstration of safety, a matter of paramount importance in the FDA’s review of investigational new drug applications for clinical trials.
Materials and Methods
Animals
The GSHPMD colony was initially housed at the University of North Carolina at Chapel Hill before being moved to Texas A&M University. Dogs were cared for and assessed according to principles outlined in the National Research Council Guide for the Care and Use of Laboratory Animals and covered by the UNC-CH Institutional Animal Care and Use Committee (IACUC) through protocols, Natural History and Immunological Parameters in the German Shorthaired Pointer Muscular Dystrophy (GSHPMD) Dog (UNC 09-011) and Standard Operating Procedures—Canine X-Linked Muscular Dystrophy (UNC 09-351 and TAMU IACUC 2015-0110).
Primer Design
Detailed primer information is available in Figure S9. Primers were designed using MacVector v14.5.0 software, and then they were checked for specificity to the region of interest of the dog genome using Primer-Blast.46 Primers were ordered from Integrated DNA Technologies. Annealing temperatures were calculated as five degrees less than the average melting temperature of the primers in a reaction. Melting temperatures were calculated using OligoAnalyzer v3.1 (IDT). Only primers that yielded the expected band size when amplified from WT dog DNA were used in our analysis.
PCR in Deletion Mapping
Genomic DNA was isolated from dog whole blood using the QIAamp DNA Blood Midi Kit (QIAGEN) following the manufacturer’s protocol. PCRs contained 0.5 μM of each primer, 200 ng genomic DNA, and 25 μL GoTaq Green Master Mix (Promega), and they were brought to a final volume of 50 μL. PCRs were performed on a PTC-200 DNA Engine thermocycler (MJ Research). Reactions were initially denatured at 95°C for 2 min and then carried through 35 cycles as follows: 95°C for 45 s, annealing for 45 s, extension at 72°C, followed by a final extension at 72°C for 10 min, and then held indefinitely at 4°C. Extension times were calculated using the processivity of standard Taq polymerase, 1 kb per minute, but reactions were extended for a minimum of 30 s. Completed PCR reactions were run on a 0.5% agarose gel, stained with ethidium bromide (Sigma-Aldrich), and captured on a digital imager (Fotodyne). PCR products from either male GSHPMD DNA or WT male GSHP DNA were compared in this manner for each primer pair.
In the case of amplifying across the deletion, reactions contained 0.5 μM primer 145 and 146, 1.5 M betaine (Sigma-Aldrich), 200 ng genomic DNA, and 25 μL GoTaq Green Master Mix (Promega), and they were brought to a final volume of 50 μL. Reactions were run on a Multigene Gradient thermocycler (Labnet International) under the following conditions: 95°C for 2 min, 35 cycles of 95°C for 30 s, 64.5°C for 30 s, 72°C for 7 min, followed by a final extension at 72°C for 10 min, and then held indefinitely at 4°C. Completed reactions were run on a 0.5% agarose gel by electrophoresis. To avoid UV-induced mutations prior to sequencing, replicate lanes were run, cut away from the gel, and stained with ethidium bromide. The position of the 2-kb band was located on the ethidium bromide-stained lanes, and then it was used to approximate the location of the band of interest on the unstained, unexposed lanes, which was excised and purified using the QIAquick Gel Extraction Kit (QIAGEN) according to the manufacturer’s protocol. The resulting DNA was used in subsequent sequencing reactions.
Assembly Gap PCR
BAC clone CH82-472H14 was ordered from the BACPAC Resource Center (CHORI). The BAC clone was provided to us in DH10B E. coli, which we propagated in LB broth (Corning Life Sciences) with 12.5 μg/mL chloramphenicol in an incubator at 37°C and 220 rpm for 20 hr. BAC DNA was purified from E. coli using the Plasmid Mini Kit (QIAGEN) following the manufacturer’s protocol.
PCRs contained 0.5 μM primer 147 and 148, 1.5 M betaine (Sigma), 5 pg BAC DNA, and 25 μL GoTaq Green Master Mix (Promega), and they were brought to a final volume of 50 μL. PCRs were performed on a PTC-200 DNA Engine thermocycler (MJ Research). Reactions were initially denatured at 95°C for 2 min and then carried through 35 cycles as follows: 95°C for 30 s, 58°C for 30 s, 72°C for 2 min, followed by a final extension at 72°C for 10 min, and then held indefinitely at 4°C. Completed reactions were run on a 0.8% agarose gel by electrophoresis, and the expected 1-kb band was excised without UV exposure or ethidium bromide staining (described in previous section) and then purified using the QIAquick Gel Extraction Kit (QIAGEN) according to the manufacturer’s protocol. The resulting DNA was used in subsequent sequencing reactions.
DNA Sequencing
The assembly gap and deletion-spanning amplicons were sequenced by the DNA Sequencing Facility at The University of Pennsylvania on a 3730xl DNA Analyzer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) according to the kit instructions. Sequencing reactions contained 8 μL Terminator Mix, 0.2 μM primer, and 100 ng gel-purified DNA in a final volume of 20 μL. Individual Sanger reads were manually trimmed, as we found software-directed trimming to be too lenient in eliminating poor-quality base calls. Trimmed reads were then assembled into a single contig using SeqMan Pro v12.0.0 (DNASTAR) under default settings. Individual Sanger reads used in our contig assembles are available in Trace Archive under the following sequential accession numbers: GSHPMD deletion reads 2,342,817,208–2,342,817,224; assembly gap reads 2,342,817,194–2,342,817,207.
Pustell Matrices, ClustalW Sequence Alignments, and Phylogenies
Pustell DNA matrices, ClustalW47 alignments, and phylogenies were created in MacVector. Pustell matrices were scored with the default DNA database matrix provided by MacVector. Specific parameters for matrices and phylogenies are provided in their respective figures. ClustalW multiple sequence alignments were performed under MacVector default conditions, which include an open gap penalty of 15.0, extend gap penalty of 6.7, and delay divergence of 30%. DNA sequences used in the alignments can be retrieved from NCBI using the coordinates provided in Figure 4B and Table S1.
tBLASTn Search
A tBLASTn search26 was performed under default conditions using the dog FTH1 peptide sequence as query against many species’ genome assemblies. Search results were exported as.csv files, which were then filtered by length and quality parameters (Figure S4). Individual hit information for each species is provided in Data S2.
Genome Assembly Builds
The following genome assembly builds were accessed via NCBI and used in our analysis (the described species are organized with common name, scientific name, genome assembly build): dog, Canis lupus familiaris, CanFam3.1; human, Homo sapiens, GRCh38.p3; chimpanzee, Pan troglodytes, Pan_troglodytes-2.1.4; mouse, Mus musculus, GRCm38.p4; cat, Felis catus, Felis_catus_8.0; pig, Sus scrofa, Sscrofa10.2; opossum, Monodelphis domestica, MonDom5; and chicken, Gallus gallus, Gallus_gallus-4.0. It should be noted that much of the gene and locus information provided herein is predicted model sequences produced by NCBI’s eukaryotic genome annotation pipeline and, therefore, subject to change due to the dynamic nature of genome assemblies and annotation software.48
Author Contributions
Conceptualization, D.J.V., P.S.H., J.N.K., H.H.S.; Methodology, D.J.V., J.N.K., H.H.S.; Investigation, D.J.V., A.S.M.; Resources, J.N.K.; Writing – Original Draft, D.J.V.; Writing – Review and Editing, A.S.M., P.S.H., J.N.K., H.H.S.; Visualization, D.J.V., A.S.M., J.N.K., H.H.S.; Supervision, P.S.H., H.H.S.; Funding Acquisition, J.N.K., H.H.S.
Acknowledgments
This work was supported by The National Institute of Neurological Disorders and Stroke (R01NS094705 and R01NS042874 to H.H.S.), The National Center for Research Resources (S10RR028027 to H.H.S.), The National Institute of Arthritis and Musculoskeletal and Skin Diseases (P30AR050950 and T32AR053461), and The Muscular Dystrophy Association (276601 to H.H.S. and 86447 to J.N.K.).
Footnotes
Supplemental Information includes nine figures, one table, and two data files and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2016.12.001.
Supplemental Information
Each species is provided in a separate sheet. Detailed information for each hit is provided, and hits remaining after each filtering criteria was applied are indicated in separate columns.
References
- 1.Ramos J., Chamberlain J.S. Gene Therapy for Duchenne muscular dystrophy. Expert Opin. Orphan Drugs. 2015;3:1255–1266. doi: 10.1517/21678707.2015.1088780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potter M.A., Chang P.L. Review--the use of immunosuppressive agents to prevent neutralizing antibodies against a transgene product. Ann. N Y Acad. Sci. 1999;875:159–174. doi: 10.1111/j.1749-6632.1999.tb08501.x. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y., Jooss K.U., Su Q., Ertl H.C., Wilson J.M. Immune responses to viral antigens versus transgene product in the elimination of recombinant adenovirus-infected hepatocytes in vivo. Gene Ther. 1996;3:137–144. [PubMed] [Google Scholar]
- 4.Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 5.Sharp N.J., Kornegay J.N., Van Camp S.D., Herbstreith M.H., Secore S.L., Kettle S., Hung W.Y., Constantinou C.D., Dykstra M.J., Roses A.D. An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy. Genomics. 1992;13:115–121. doi: 10.1016/0888-7543(92)90210-j. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman E.P., Morgan J.E., Watkins S.C., Partridge T.A. Somatic reversion/suppression of the mouse mdx phenotype in vivo. J. Neurol. Sci. 1990;99:9–25. doi: 10.1016/0022-510x(90)90195-s. [DOI] [PubMed] [Google Scholar]
- 7.Pigozzo S.R., Da Re L., Romualdi C., Mazzara P.G., Galletta E., Fletcher S., Wilton S.D., Vitiello L. Revertant fibers in the mdx murine model of Duchenne muscular dystrophy: an age- and muscle-related reappraisal. PLoS ONE. 2013;8:e72147. doi: 10.1371/journal.pone.0072147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schatzberg S.J., Anderson L.V., Wilton S.D., Kornegay J.N., Mann C.J., Solomon G.G., Sharp N.J. Alternative dystrophin gene transcripts in golden retriever muscular dystrophy. Muscle Nerve. 1998;21:991–998. doi: 10.1002/(sici)1097-4598(199808)21:8<991::aid-mus2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Flanigan K.M., Campbell K., Viollet L., Wang W., Gomez A.M., Walker C.M., Mendell J.R. Anti-dystrophin T cell responses in Duchenne muscular dystrophy: prevalence and a glucocorticoid treatment effect. Hum. Gene Ther. 2013;24:797–806. doi: 10.1089/hum.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olby N.J., Sharp N.J., Nghiem P.E., Keene B.W., DeFrancesco T.C., Sidley J.A., Kornegay J.N., Schatzberg S.J. Clinical progression of X-linked muscular dystrophy in two German Shorthaired Pointers. J. Am. Vet. Med. Assoc. 2011;238:207–212. doi: 10.2460/javma.238.2.207. [DOI] [PubMed] [Google Scholar]
- 11.Schatzberg S.J., Olby N.J., Breen M., Anderson L.V., Langford C.F., Dickens H.F., Wilton S.D., Zeiss C.J., Binns M.M., Kornegay J.N. Molecular analysis of a spontaneous dystrophin ‘knockout’ dog. Neuromuscul. Disord. 1999;9:289–295. doi: 10.1016/s0960-8966(99)00011-5. [DOI] [PubMed] [Google Scholar]
- 12.Brown J., Dry K.L., Edgar A.J., Pryde F.E., Hardwick L.J., Aldred M.A., Lester D.H., Boyle S., Kaplan J., Dufier J.L. Analysis of three deletion breakpoints in Xp21.1 and the further localization of RP3. Genomics. 1996;37:200–210. doi: 10.1006/geno.1996.0543. [DOI] [PubMed] [Google Scholar]
- 13.Francke U., Ochs H.D., de Martinville B., Giacalone J., Lindgren V., Distèche C., Pagon R.A., Hofker M.H., van Ommen G.J., Pearson P.L. Minor Xp21 chromosome deletion in a male associated with expression of Duchenne muscular dystrophy, chronic granulomatous disease, retinitis pigmentosa, and McLeod syndrome. Am. J. Hum. Genet. 1985;37:250–267. [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel L.M., Monaco A.P., Middlesworth W., Ochs H.D., Latt S.A. Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc. Natl. Acad. Sci. USA. 1985;82:4778–4782. doi: 10.1073/pnas.82.14.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monaco A.P., Neve R.L., Colletti-Feener C., Bertelson C.J., Kurnit D.M., Kunkel L.M. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 16.Smith T.J., Wilson L., Kenwrick S.J., Forrest S.M., Speer A., Coutelle C., Davies K.E. Isolation of a conserved sequence deleted in Duchenne muscular dystrophy patients. Nucleic Acids Res. 1987;15:2167–2174. doi: 10.1093/nar/15.5.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benton M.J., Donoghue P.C. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 18.Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., 3rd, Zody M.C. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 19.Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald M., Shenk T. The sequence 5’-AAUAAA-3’forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981;24:251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- 21.Proudfoot N.J., Brownlee G.G. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 22.Tabaska J.E., Zhang M.Q. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- 23.Andrews S.C. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta. 2010;1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Costanzo F., Colombo M., Staempfli S., Santoro C., Marone M., Frank R., Delius H., Cortese R. Structure of gene and pseudogenes of human apoferritin H. Nucleic Acids Res. 1986;14:721–736. doi: 10.1093/nar/14.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Harrison P.M., Liu Y., Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christophe-Hobertus C., Szpirer C., Guyon R., Christophe D. Identification of the gene encoding Brain Cell Membrane Protein 1 (BCMP1), a putative four-transmembrane protein distantly related to the Peripheral Myelin Protein 22 / Epithelial Membrane Proteins and the Claudins. BMC Genomics. 2001;2:3. doi: 10.1186/1471-2164-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christophe-Hobertus C., Kooy F., Gecz J., Abramowicz M.J., Holinski-Feder E., Schwartz C., Christophe D. TM4SF10 gene sequencing in XLMR patients identifies common polymorphisms but no disease-associated mutation. BMC Med. Genet. 2004;5:22. doi: 10.1186/1471-2350-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J.M., Cooper D.N., Chuzhanova N., Férec C., Patrinos G.P. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 30.Axelsson E., Webster M.T., Ratnakumar A., Ponting C.P., Lindblad-Toh K., LUPA Consortium Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S., Lee S.H., Chung T.G., Kim H.J., Yoon T.K., Kwak I.P., Park S.H., Cha W.T., Cho S.W., Cha K.Y. Molecular and cytogenetic characterization of two azoospermic patients with X-autosome translocation. J. Assist. Reprod. Genet. 2003;20:385–389. doi: 10.1023/A:1025437329427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei M., Rooney A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown D.D., Wensink P.C., Jordan E. A comparison of the ribosomal DNA’s of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J. Mol. Biol. 1972;63:57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- 34.Haber J.E., Ira G., Malkova A., Sugawara N. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday’s model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:79–86. doi: 10.1098/rstb.2003.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta P.K., Adamtziki E., Budde U., Jaiprakash M., Kumar H., Harbeck-Seu A., Kannan M., Oyen F., Obser T., Wedekind I. Gene conversions are a common cause of von Willebrand disease. Br. J. Haematol. 2005;130:752–758. doi: 10.1111/j.1365-2141.2005.05660.x. [DOI] [PubMed] [Google Scholar]
- 36.Pink R.C., Wicks K., Caley D.P., Punch E.K., Jacobs L., Carter D.R. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi S., Fujihara Y., Mise N., Kaseda K., Abe K., Ishino F., Okabe M. The X-linked imprinted gene family Fthl17 shows predominantly female expression following the two-cell stage in mouse embryos. Nucleic Acids Res. 2010;38:3672–3681. doi: 10.1093/nar/gkq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P.J., McCarrey J.R., Yang F., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 39.Ruzzenenti P., Asperti M., Mitola S., Crescini E., Maccarinelli F., Gryzik M., Regoni M., Finazzi D., Arosio P., Poli M. The Ferritin-Heavy-Polypeptide-Like-17 (FTHL17) gene encodes a ferritin with low stability and no ferroxidase activity and with a partial nuclear localization. Biochim. Biophys. Acta. 2015;1850:1267–1273. doi: 10.1016/j.bbagen.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 41.Hoeppner M.P., Lundquist A., Pirun M., Meadows J.R., Zamani N., Johnson J., Sundström G., Cook A., FitzGerald M.G., Swofford R. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE. 2014;9:e91172. doi: 10.1371/journal.pone.0091172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graumann M.B., DeRose S.A., Ostrander E.A., Storb R. Polymorphism analysis of four canine MHC class I genes. Tissue Antigens. 1998;51:374–381. doi: 10.1111/j.1399-0039.1998.tb02976.x. [DOI] [PubMed] [Google Scholar]
- 43.Kennedy L.J., Barnes A., Happ G.M., Quinnell R.J., Courtenay O., Carter S.D., Ollier W.E., Thomson W. Evidence for extensive DLA polymorphism in different dog populations. Tissue Antigens. 2002;60:43–52. doi: 10.1034/j.1399-0039.2002.600106.x. [DOI] [PubMed] [Google Scholar]
- 44.Ross P., Buntzman A.S., Vincent B.G., Grover E.N., Gojanovich G.S., Collins E.J., Frelinger J.A., Hess P.R. Allelic diversity at the DLA-88 locus in Golden Retriever and Boxer breeds is limited. Tissue Antigens. 2012;80:175–183. doi: 10.1111/j.1399-0039.2012.01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 46.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruitt K.D., Tatusova T., Brown G.R., Maglott D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each species is provided in a separate sheet. Detailed information for each hit is provided, and hits remaining after each filtering criteria was applied are indicated in separate columns.