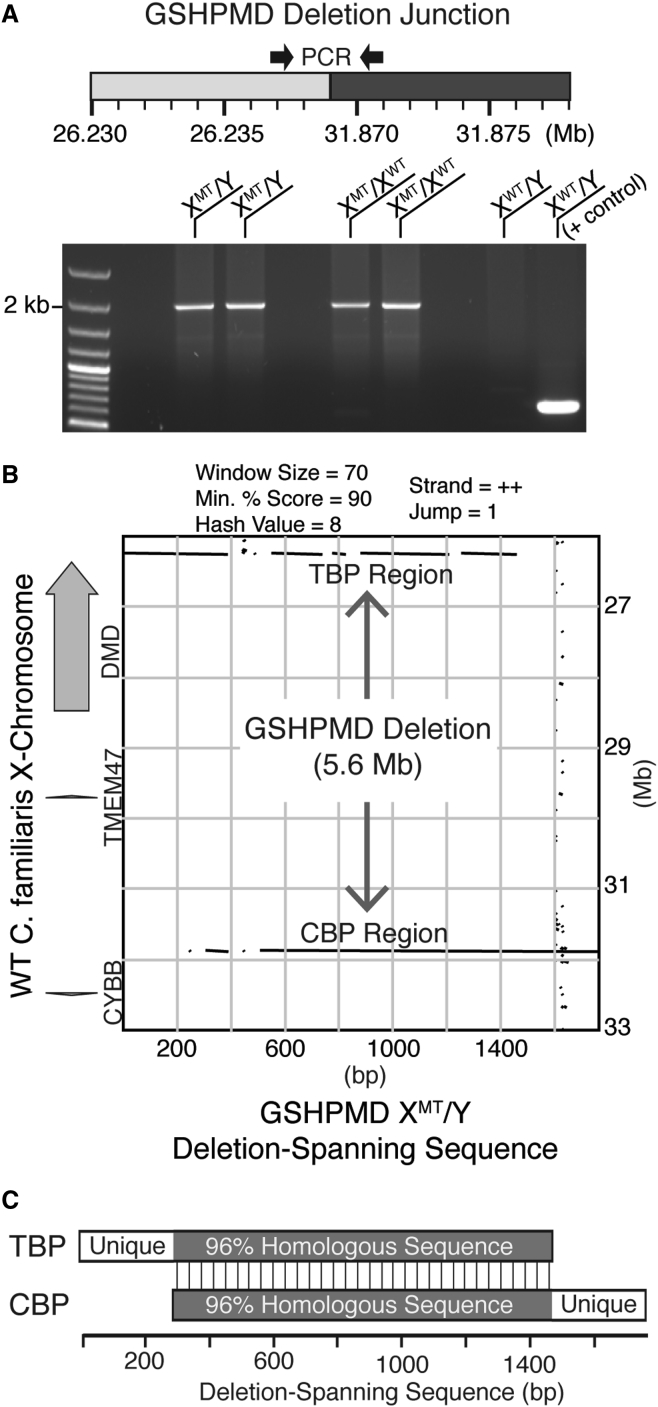
Figure 3.
PCR Amplification across the 5.6-Mb Deletion in the GSHPMD Model
(A) Schematic showing primers that are spaced over 5.6 Mb apart in wild-type dog. PCR products generated from this primer pair using DNA from two affected males, two carrier females, and one wild-type male are displayed on an agarose gel following electrophoresis. A positive control using an unrelated primer pair is provided in the rightmost lane for the wild-type dog. (B) Pustell DNA matrix comparing the sequenced deletion-spanning amplicon from a GSHPMD male to the indicated region of wild-type dog X chromosome. Black lines indicate homology between the compared sequences. (C) Macroscale DNA sequence comparison of the telomeric and centromeric deletion breakpoints. Shaded region indicates >96% homology between the deletion breakpoints. XMT/Y, affected male; XMT/XWT, carrier female; XWT/Y, wild-type male; TBP, telomeric breakpoint; CBP, centromeric breakpoint; WT, wild-type.