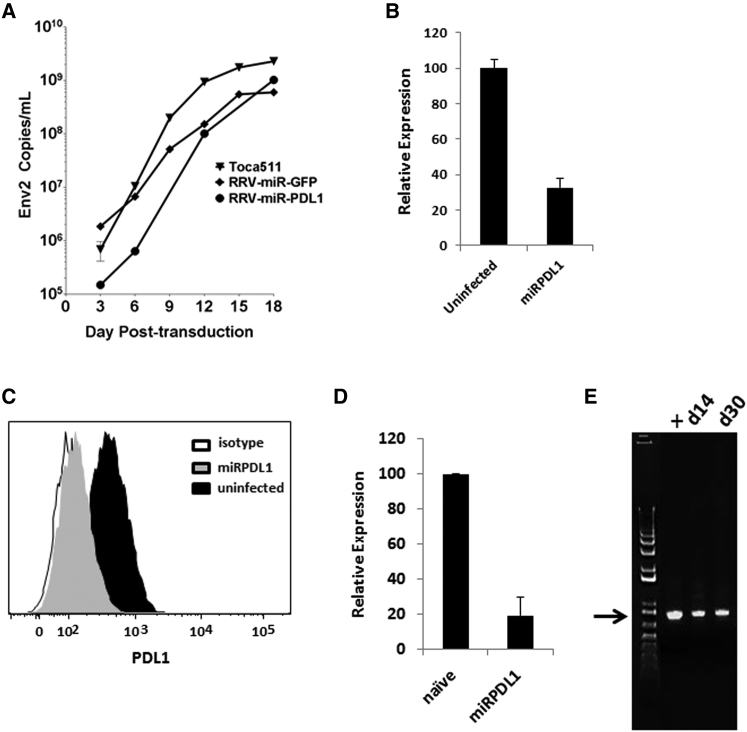
Figure 3.
Vector Characterization of RRVs Expressing miRshRNA
(A) Replication kinetics of RRV-miRGFP and RRV-miRPDL1. The viral genome in the supernatants of infected LN-18 cells (MOI of 0.1) at indicated time points were quantified by qRT-PCR using primer set targeted to the env region (Figure 1). RRV-yCD2 was included as a positive control. A paired t test was performed and showed no statistically significant difference in replication kinetics between RRV-miRGFP versus Toca 511 (p = 0.0778) and between RRV-miRPDL1 versus Toca 511 (p = 0.1915). (B) PDL1 gene expression in RRV-miRPDL1 infected LN-18 cells relative to uninfected cells. The values presented are means ± SD of triplicates. (C) LN-18 cells infected with RRV-miRPDL1 were stained for PDL1 cell surface expression with PDL1 antibody and analyzed by flow cytometry. The histogram shown represents data from one of the three independent experiments. (D) PDL1 cell surface expression in RRV-miRPDL1 infected LN-18 cells relative to uninfected cells. The values presented are means ± SD of three independent experiments. (E) Vector stability of RRV-miRPDL1 in LN-18 cells was analyzed by endpoint PCR at 14 and 30 days post infection. The DNA molecular marker (1 Kb Plus marker, Invitrogen) is included in the first lane. positive control using plasmid DNA as the template, +. The arrow indicates the expected size of the PCR products (844 bp).