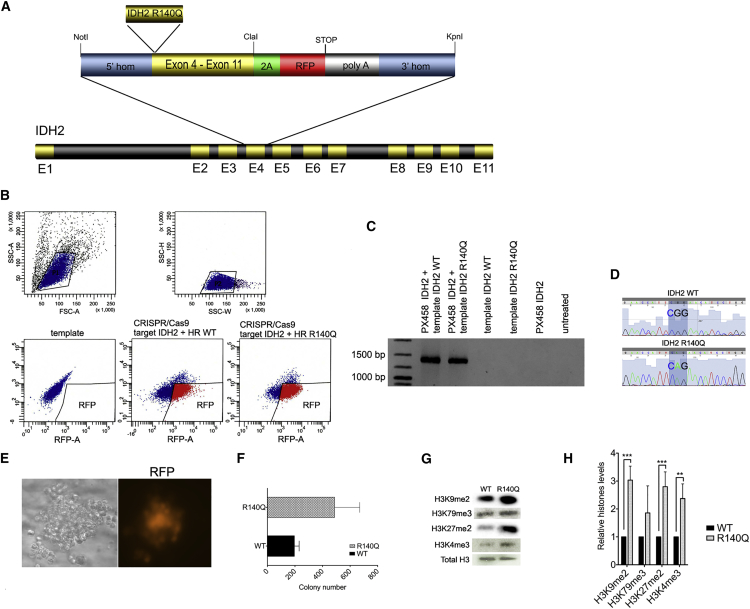
Figure 1.
CRISPR-Cas9-Mediated Introduction of IDH2 R140Q Mutation in Human K562 Leukemia Cells
(A) Schematic of the DNA repair template for introduction of IDH2 R140Q mutation into human K562 cells via homologous recombination. (B) FACS sorting of K562 cells after transfection with the CRISPR-Cas9 plasmid and plasmid carrying the DNA template for HR; upper left: forward scatter (FSC); upper right: side scatter (SSC); lower left: K562 cells transfected with the plasmid carrying the DNA template only; lower middle: K562 cells transfected with the plasmid carrying the DNA template, including the wt IDH2 sequence plus CRIPSR-Cas9/sgRNA; lower right: K562 cells transfected with the plasmid carrying the DNA template, including the mutant IDH2 sequence plus CRIPSR-Cas9/sgRNA. (C) Targeted integration (TI) PCR confirming introduction of the DNA templates, including the IDH2 R140Q mutation or wt IDH2 sequence, respectively, at the IDH2 locus. (D) Sanger DNA sequences confirming accurate introduction of the IDH2 R140Q mutation (and IDH2 wt sequence) at the IDH2 gene locus. (E) Colony of human K562 cells plated into methylcellulose after FACS sorting for RFP expression; left: colony morphology in brightfield microscopy; right: RFP detection via fluorescence microscopy. (F) Histograph representing colony formation of K562 cells after successful genome editing with introduction of the IDH2 R140Q mutation and re-introduction of the IDH2 wt sequence, respectively, and after 7 days post-FACS sorting. Error bars indicate SD of three independent experiments. (G) Immunoblotting of purified histone extracts from K562 IDH2 wt and K562 IDH2 R140Q. (H) Bar chart representing quantification of specific histone levels relative to total histone 3. Error bars indicate SD of three independent experiments (±SD, **p < 0.01, ***p < 0.001).