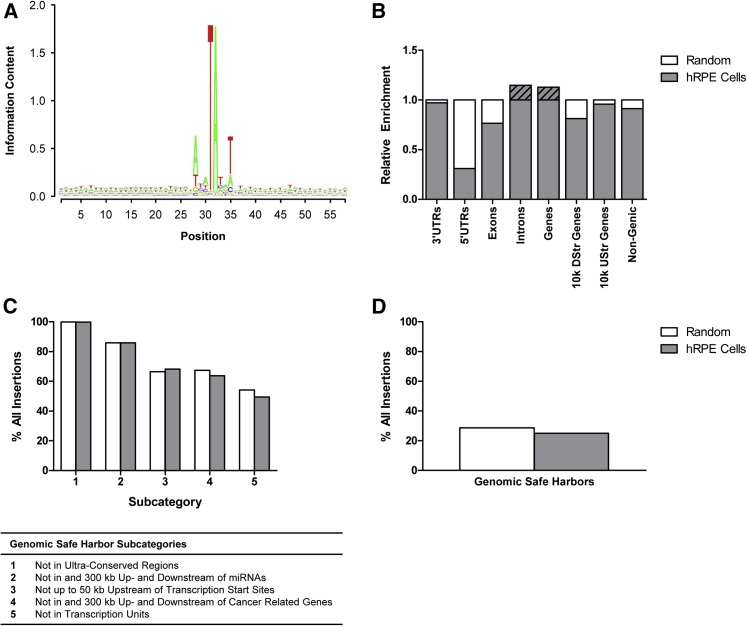
Figure 7.
Integration Profiling of the pFAR4-ITRs CMV PEDF BGH Transposon in Human RPE Cells
(A) Consensus sequence of the Sleeping Beauty insertion sites. The y axis in the Seqlogo analysis represents the strength of the information, with two bits being the maximum for a DNA sequence. (B) SB integration sites in human RPE cells were compared to a random control set of 1 × 104 computationally generated loci, determining the frequencies of integrations into annotated genomic features including 3′ UTRs, 5′ UTRs, exons, introns, gene bodies, and regions around transcription start sites. The integration profile was found to be close-to-random. (C) Relative frequencies of SB insertions into chromosomal sites defined as genomic safe harbors in comparison with the random dataset. (D) Relative frequency of SB integrations into genomic safe harbors, cumulative for all five subcategories shown in (C), in comparison with the random dataset.