Figure 4. A statistical representation of the deleterious/damaging nsSNP predicted by various insilico tools.
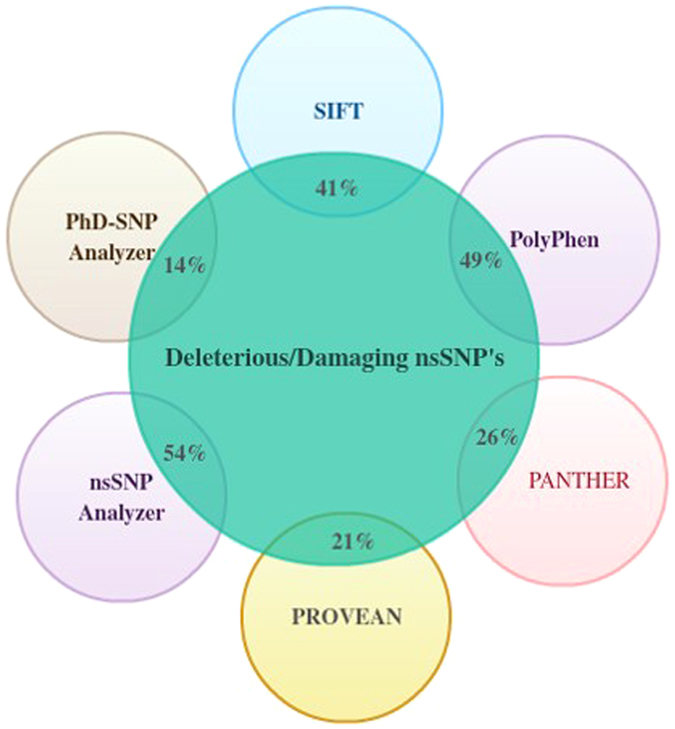
Percent deleterious nsSNP’s predicted by SIFT, PolyPhen, PANTHER, PROVEAN, nsSNPAnalyzer, PhD-SNP Analyzer. Out of 95 HOXB13 non-homeobox nsSNP’s SIFT predicted 41%, PolyPhen predicted 49%, PANTHER predicted 26%, PROVEAN 21%, nsSNPAnalyzer 54% and PhD-SNPAnalyzer 14% of the nsSNP’s to be potentially deleterious/damaging.
