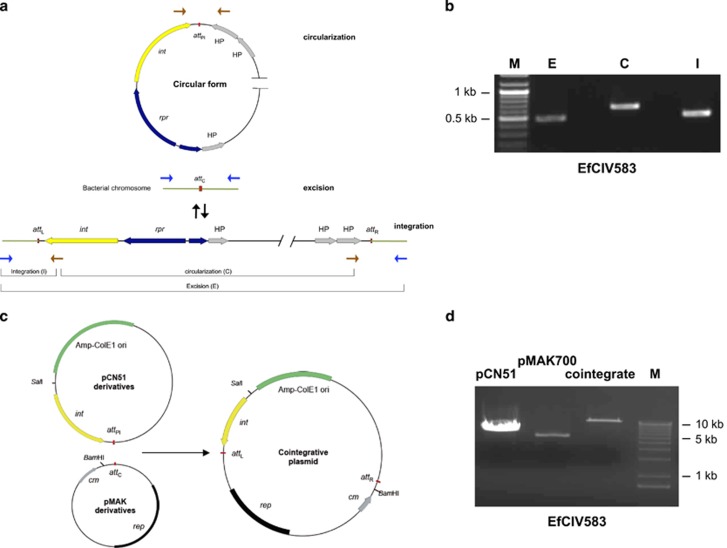
Figure 2.
Characterization of EfCIV583-encoded Int protein. (a) Schematic representation of the EfCIV583-dependent excision and circularization processes. The relevant genes, genetic markers and PCR primer binding sites are shown. (b) Detection of EfCIV583 excision and circularization. DNA from E. faecalis VE14089 was extracted and PCR-amplified using specific primers (see scheme in a) recognizing the external and internal sequence of the element (integration: I), primers recognizing the flanking sequences (excision: E) or PCR-amplified using a pair of primers set divergently at both termini of the island (circularization: C). M: molecular weight marker. (c) Constructs used for test site-specific integration in E. coli. Top: pCN51 derivatives containing the EfCIV583 attPI-int gene. Bottom: thermosensitive derivatives of pMAK700 carrying the attC from the E. faecalis chromosome. The relevant genetic markers and restriction enzyme sites are shown. (d) Plasmid DNA was isolated from overnight cultures grown at 37 °C (for strains carrying uniquely the pCN51 or pMAK700 derivatives) or at 43 °C (for strain carrying both plasmids), in the presence of ampicillin (pCN51) or chloramphenicol (pMAK700 derivative and cointegrative plasmid). Plasmids were digested with BamHI (pMAK700 derivatives) or with SalI (pCN51 derivatives and cointegrative plasmids).