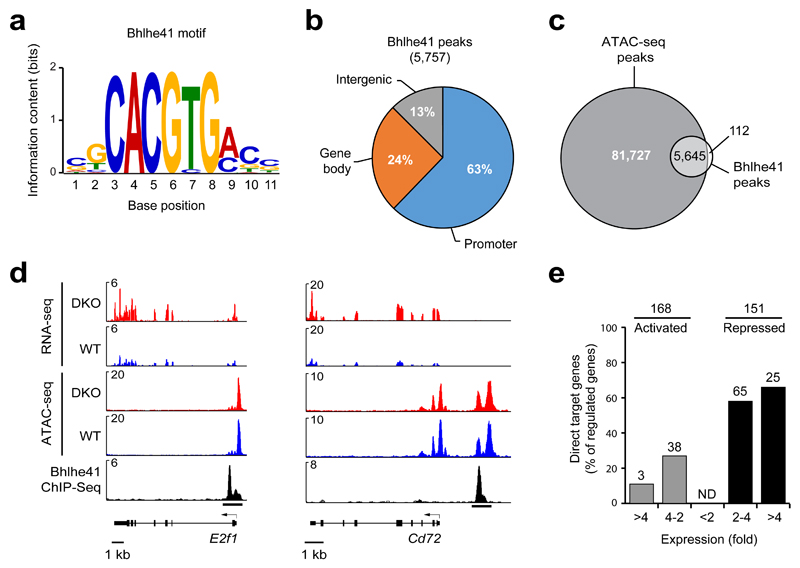
Figure 5. Identification of regulated Bhlhe41 target genes in B-1a cells.
(a) Consensus Bhlhe41-binding motif identified with an E-value of 1.3 x 10–120 by the de novo motif-discovery program MEME-ChIP. (b) Presence of Bhlhe41 peaks at the indicated gene regions. (c) Overlap of Bhlhe41 peaks with open chromatin regions, which were mapped by ATAC-seq39. (d) Presence of open chromatin (ATAC-seq) and RNA transcripts (RNA-seq) at the repressed Bhlhe41 target genes E2f1 and Cd72 in VH12/Vκ4 transgenic wild-type and DKO B-1a cells. Bhlhe41 binding was determined by ChIP-seq analysis of VH12/Vκ4 transgenic Bhlhe41Tag/Tag B-1a cells. (e) Identification of regulated Bhlhe41 target genes in VH12/Vκ4 transgenic B-1a cell. The number (above bars) and frequency (vertical axis) of Bhlhe41 target genes is shown together with the indicated differences in mRNA expression (horizontal axis) in VH12/Vκ4 transgenic DKO B-1a cells relative to their expression in VH12/Vκ4 transgenic wild-type B-1a cells. The 168 activated and 151 repressed genes (without considering Bhlhe41 binding) were selected for an adjusted P value of < 0.05 and a TPM value (transcripts per million) of > 5 in one of the two cell types.