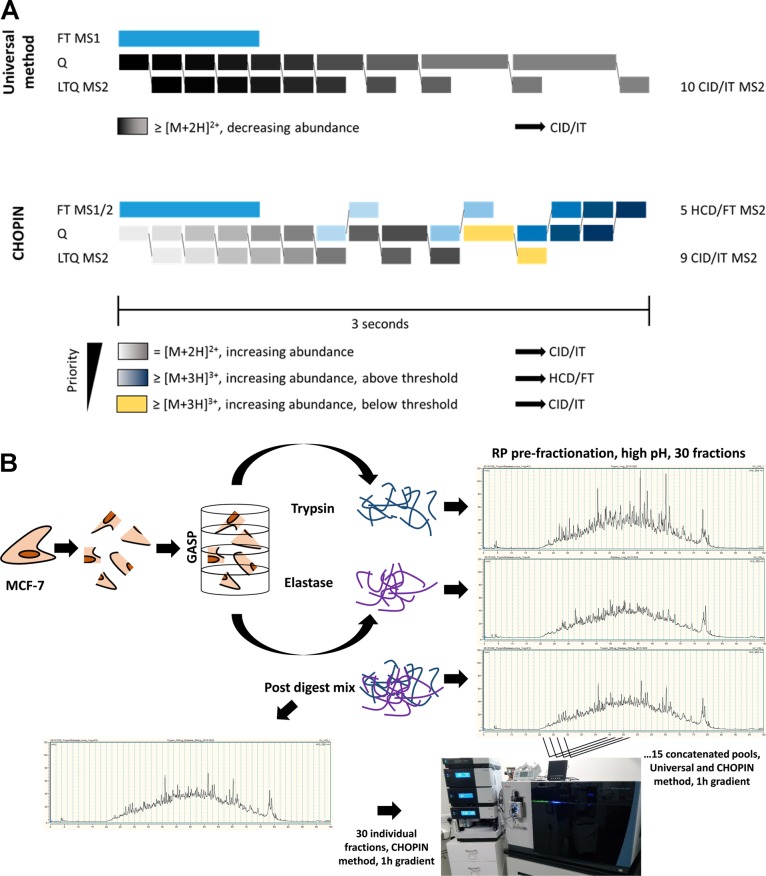
Figure 1.
Comprehensive cell proteome coverage by prefractionation and CHOPIN MS analysis workflow. (A) Mass spectrometry acquisition methods demonstrating the dynamic segmentation of analytical channels for MS1 FT (Orbitrap), Q (Quadrupole), and MS2 LTQ (Linear Ion Trap) that were designed for the Universal (upper panel) and CHarge Ordered Parallel Ion aNalysis (CHOPIN) method (lower panel). The Universal Method makes use of the parallel acquisition of MS1 scan in the Orbitrap, while peptide fragments are scanned in the LTQ, ordered by decreasing precursor intensity. Additional parallelization is achieved by concurrent MS2 scans and isolation of the following precursor. Precursor ion accumulation is allowed to proceed for up to 250 ms if no previously unselected precursor is found. CHOPIN adds another level of parallelization by triaging intense and highly charged ions to be analyzed by an Orbitrap MS2 scan, while low abundant precursor ions are prioritized for the more sensitive MS2 scan in the linear ion trap. CHOPIN and the data analysis is further described in Supporting Information. (B) Methodological workflow for the analysis of the MCF-7 breast cancer cell line deep proteome. MCF-7 cell extracts were digested with either trypsin or elastase, and peptide mixtures were separated by high-pH reversed-phase (RP) HPLC to collect 30 fractions that were pooled in a concatenated fashion to 15 fractions. Also, tryptic and elastase digest was mixed and prefractionated as above (“Post Digest Mix”, PDM), followed by concatenation or distinct fraction analysis. Each fraction was subsequently analyzed by LC–MS/MS using both the Universal and CHOPIN acquisition methods. Detailed results for each individual experiment are shown in the Supporting Information. (Orbitrap Fusion Lumos photo by RF).