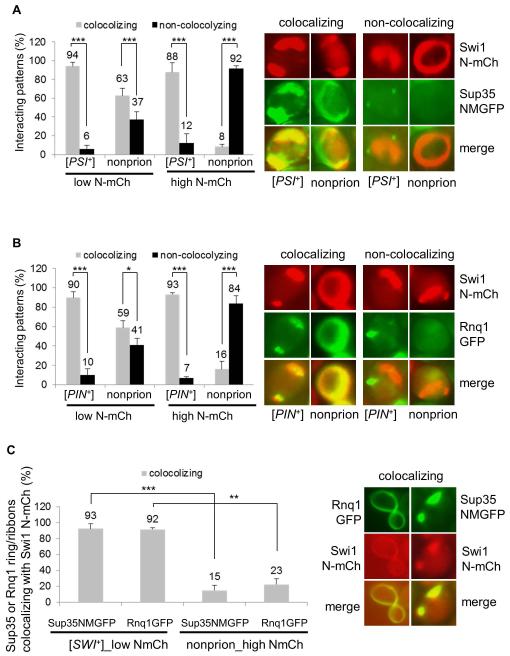
Figure 6. Interacting mechanisms of heterologous prion proteins, Sup35, Rnq1 and Swi1, in their prionogenesis.
(A) The indicated 74D-694 strains were co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP. The ring/ribbon-like Swi1 N-mCherry aggregates (NmCh) that are supposed to be prionogenic in Swi1 prionogenesis were quantified for their interaction with Sup35-NMGFP (NMGFP) at low (low N-mCh, 0.02% galactose) or high (high N-mCh, 0.5% galactose) production of Swi1 PrD. Notably, 10 μM CuSO4 was supplemented, and both co-localizing and non-co-localizing patterns were observed after 48 h induction. The images shown on the right are representatives and plots are a summary of data from at least three independent experiments. (B) Similar to panel A, but the 74D-694 strains were co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP, and the interaction is for Swi1 N-mCh and Rnq1-GFP. (C) A [SWI+] or a non-prion 74D-694 strain was co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP, or with p423GAL1-NmCherry pCUP1-RNQ1GFP. Subsequently, the newly formed Sup35 NM-GFP (NMGFP) and Rnq1-GFP ring/ribbon-like aggregates were assayed for colocalization with Swi1 N-mCherry after 48 h of incubation. Low NmCh, 0.02% galactose; high NmCh, 0.5% galactose. In the experiment, 10 μM CuSO4 was supplied as minimum to visualize Sup35 NMGFP and Rnq1-GFP aggregation. Here shows a summary of the colocalizing frequencies (left) and representative images of the co-localization (right). Statistical analysis in the figure was performed by T test (*, P<0.05; **, P<0.01; ***, P<0.001).